Post suggestions and read about updates in the Comments section.
![image]() This tutorial introduces researchers to considerations in somatic short variant discovery using GATK4 Mutect2. Example data are based on a breast cancer cell line and its matched normal cell line derived from blood and are aligned to GRCh38 with post-alt processing [1]. The tutorial focuses on how to call traditional somatic short mutations, as described in Article#11127 and pipelined in GATK v4.0.0.0's mutect2.wdl [2]. The tool and its workflow are in BETA status as of this writing, which means they may undergo changes and are not guaranteed for production.
This tutorial introduces researchers to considerations in somatic short variant discovery using GATK4 Mutect2. Example data are based on a breast cancer cell line and its matched normal cell line derived from blood and are aligned to GRCh38 with post-alt processing [1]. The tutorial focuses on how to call traditional somatic short mutations, as described in Article#11127 and pipelined in GATK v4.0.0.0's mutect2.wdl [2]. The tool and its workflow are in BETA status as of this writing, which means they may undergo changes and are not guaranteed for production.
► For Broad Mutation Calling Best Practices, see FireCloud Article#45055.
Section 1 calls somatic mutations with Mutect2 using all the bells and whistles of the tool. Section 2 outlines how to create the panel of normals resource using the tumor-only mode of Mutect2. Section 3 outlines how to estimate cross-sample contamination. Section 4 shows how to filter the callset with FilterMutectCalls. Unlike GATK3, in GATK4 the somatic calling and filtering functionalities are embodied by separate tools. Section 5 shows an optional filtering step to filter by sequence context artifacts that present with orientation bias, e.g. OxoG artifacts. Section 6 shows how to set up in IGV for manual review. Finally, section 7 provides a brief list of related resources that may be of interest to researchers.
GATK4 Mutect2 is a versatile variant caller that not only is more sensitive than, but is also roughly twice as fast as, HaplotypeCaller's reference confidence mode. Researchers who wish to customize analyses should find the tutorial's descriptions of the multiple levers of Mutect2 in section 1 and descriptions of the tumor-only mode of Mutect2 in section 2 of interest.
Jump to a section
- Call somatic short variants and generate a bamout with Mutect2
☞ 1.1 What are the Mutect2 annotations?
☞ 1.2 What is the impact of disabling the MateOnSameContigOrNoMappedMateReadFilter read filter?
- Create a sites-only PoN with CreateSomaticPanelOfNormals
☞ 2.1 The tumor-only mode of Mutect2 is useful outside of pon creation
- Estimate cross-sample contamination using GetPileupSummaries and CalculateContamination
☞ 3.1 What if I find high levels of contamination?
- Filter for confident somatic calls using FilterMutectCalls
- (Optional) Estimate artifacts with CollectSequencingArtifactMetrics and filter them with FilterByOrientationBias
☞ 5.1 Tally of applied filters for the tutorial data
- Set up in IGV to review somatic calls
- Related resources
Tools involved
- GATK v4.0.0.0 is available in a Docker image and as a standalone jar. For the latest release, see the Downloads page. Note that GATK v4.0.0.0 contains Picard tools from release v2.17.2 that are callable with the
gatk launch script.
- Desktop IGV. The tutorial uses v2.3.97.
Download example data
Download tutorial_11136.tar.gz, either from the GoogleDrive or from the ftp site. To access the ftp site, leave the password field blank. If the GoogleDrive link is broken, please let us know. The tutorial also requires the GRCh38 reference FASTA, dictionary and index. These are available from the GATK Resource Bundle. For details on the example data and resources, see [3] and [4].
► The tutorial steps switch between the subset and full data. Some of the data files, e.g. BAMs, are restricted to a small region of the genome to efficiently pace the tutorial. Other files, e.g. the Mutect2 calls that the tutorial filters, are from the entire genome. The tutorial content was originally developed for the 2017-09 Helsinki workshop and we make the full data files, i.e. the resource files and the BAMs, available at gs://gatk-best-practices/somatic-hg38.
1. Call somatic short variants and generate a bamout with Mutect2
Here we have a rather complex command to call somatic variants on the HCC1143 tumor sample using Mutect2. For a synopsis of what somatic calling entails, see Article#11127. The command calls somatic variants in the tumor sample and uses a matched normal, a panel of normals (PoN) and a population germline variant resource.
gatk --java-options "-Xmx2g" Mutect2 \
-R hg38/Homo_sapiens_assembly38.fasta \
-I tumor.bam \
-I normal.bam \
-tumor HCC1143_tumor \
-normal HCC1143_normal \
-pon resources/chr17_pon.vcf.gz \
--germline-resource resources/chr17_af-only-gnomad_grch38.vcf.gz \
--af-of-alleles-not-in-resource 0.0000025 \
--disable-read-filter MateOnSameContigOrNoMappedMateReadFilter \
-L chr17plus.interval_list \
-O 1_somatic_m2.vcf.gz \
-bamout 2_tumor_normal_m2.bam
This produces a raw unfiltered somatic callset 1_somatic_m2.vcf.gz, a reassembled reads BAM 2_tumor_normal_m2.bam and the respective indices 1_somatic_m2.vcf.gz.tbi and 2_tumor_normal_m2.bai.
Comments on select parameters
- Specify the case sample for somatic calling with two parameters. Provide the BAM with
-I and the sample's read group sample name (the SM field value) with -tumor. To look up the read group SM field use GetSampleName. Alternatively, use samtools view -H tumor.bam | grep '@RG'.
- Prefilter variant sites in a control sample alignment. Specify the control BAM with
-I and the control sample's read group sample name (the SM field value) with -normal. In the case of a tumor with a matched normal control, we can exclude even rare germline variants and individual-specific artifacts. If we analyze our tumor sample with Mutect2 without the matched normal, we get an order of magnitude more calls than with the matched normal.
- Prefilter variant sites in a panel of normals callset. Specify the panel of normals (PoN) VCF with
-pon. Section 2 outlines how to create a PoN. The panel of normals not only represents common germline variant sites, it presents commonly noisy sites in sequencing data, e.g. mapping artifacts or other somewhat random but systematic artifacts of sequencing. By default, the tool does not reassemble nor emit variant sites that match identically to a PoN variant. To enable genotyping of PoN sites, use the --genotype-pon-sites option. If the match is not exact, e.g. there is an allele-mismatch, the tool reassembles the region, emits the calls and annotates matches in the INFO field with IN_PON.
- Annotate variant alleles by specifying a population germline resource with
--germline-resource. The germline resource must contain allele-specific frequencies, i.e. it must contain the AF annotation in the INFO field [4]. The tool annotates variant alleles with the population allele frequencies. When using a population germline resource, consider adjusting the --af-of-alleles-not-in-resource parameter from its default of 0.001. For example, the gnomAD resource af-only-gnomad_grch38.vcf.gz represents ~200k exomes and ~16k genomes and the tutorial data is exome data, so we adjust --af-of-alleles-not-in-resource to 0.0000025 which corresponds to 1/(2*exome samples). The default of 0.001 is appropriate for human sample analyses without any population resource. It is based on the human average rate of heterozygosity. The population allele frequencies (POP_AF) and the af-of-alleles-not-in-resource factor in probability calculations of the variant being somatic.
- Include reads whose mate maps to a different contig. For our somatic analysis that uses alt-aware and post-alt processed alignments to GRCh38, we disable a specific read filter with
--disable-read-filter MateOnSameContigOrNoMappedMateReadFilter. This filter removes from analysis paired reads whose mate maps to a different contig. Because of the way BWA crisscrosses mate information for mates that align better to alternate contigs (in alt-aware mapping to GRCh38), we want to include these types of reads in our analysis. Otherwise, we may miss out on detecting SNVs and indels associated with alternate haplotypes. Disabling this filter deviates from current production practices.
- Target the analysis to specific genomic intervals with the
-L parameter. Here we specify this option to speed up our run on the small tutorial data. For the full callset we use in section 4, calling was on the entirety of the data, without an intervals file.
- Generate the reassembled alignments file with
-bamout. The bamout alignments contain the artificial haplotypes and reassembled alignments for the normal and tumor and enable manual review of calls. The parameter is not required by the tool but is recommended as adding it costs only a small fraction of the total run time.
To illustrate how Mutect2 applies annotations, below are five multiallelic sites from the full callset. Pull these out with gzcat somatic_m2.vcf.gz | awk '$5 ~","'. The awk '$5 ~","' subsets records that contain a comma in the 5th column.
We see eleven columns of information per variant call including genotype calls for the normal and tumor. Notice the empty fields for QUAL and FILTER, and annotations at the site (INFO) and sample level (columns 10 and 11). The samples each have genotypes and when a site is multiallelic, we see allele-specific annotations. Samples may have additional annotations, e.g. PGT and PID that relate to phasing.
☞ 1.1 What are the Mutect2 annotations?
We can view the standard FORMAT-level and INFO-level Mutect2 annotations in the VCF header.
The Variant Annotations section of the Tool Documentation further describe some of the annotations. For a complete list of annotations available in GATK4, see this site.
To enable specific filtering that relies on nonstandard annotations, or just to add additional annotations, use the -A argument. For example, -A ReferenceBases adds the ReferenceBases annotation to variant calls. Note that if an annotation a filter relies on is absent, FilterMutectCalls will skip the particular filtering without any warning messages.
☞ 1.2 What is the impact of disabling the MateOnSameContigOrNoMappedMateReadFilter read filter?
To understand the impact, consider some numbers. After all other read filters, the MateOnSameContigOrNoMappedMateReadFilter (MOSCO) filter additionally removes from analysis 8.71% (8,681,271) tumor sample reads and 8.18% (6,256,996) normal sample reads from the full data. The impact of disabling the MOSCO filter is that reads on alternate contigs and read pairs that span contigs can now lend support to variant calls.
For the tutorial data, including reads normally filtered by the MOSCO filter roughly doubles the number of Mutect2 calls. The majority of the additional calls comes from the ALT, HLA and decoy contigs.
back to top
2. Create a sites-only PoN with CreateSomaticPanelOfNormals
We make the motions of creating a PoN using three germline samples. These samples are HG00190, NA19771 and HG02759 [3].
First, run Mutect2 in tumor-only mode on each normal sample. In tumor-only mode, a single case sample is analyzed with the -tumor flag without an accompanying matched control -normal sample. For the tutorial, we run this command only for sample HG00190.
gatk Mutect2 \
-R ~/Documents/ref/hg38/Homo_sapiens_assembly38.fasta \
-I HG00190.bam \
-tumor HG00190 \
--disable-read-filter MateOnSameContigOrNoMappedMateReadFilter \
-L chr17plus.interval_list \
-O 3_HG00190.vcf.gz
This generates a callset 3_HG00190.vcf.gz and a matching index. Mutect2 calls variants in the sample with the same sensitive criteria it uses for calling mutations in the tumor in somatic mode. Because the command omits the use of options that trigger upfront filtering, we expect all detectable variants to be called. The calls will include low allele fraction variants and sites with multiple variant alleles, i.e. multiallelic sites. Here are two multiallelic records from 3_HG00190.vcf.gz.
We see for each site, Mutect2 calls the ref allele and three alternate alleles. The GT genotype call is 0/1/2/3. The AD allele depths are 16,3,12,4 and 41,5,24,4, respectively for the two sites.
Comments on select parameters
- One option that is not used here is to include a germline resource with
--germline-resource. Remember from section 1 this resource must contain AF population allele frequencies in the INFO column. Use of this resource in tumor-only mode, just as in somatic mode, allows upfront filtering of common germline variant alleles. This effectively omits common germline variant alleles from the PoN. Note the related optional parameter --max-population-af (default 0.01) defines the cutoff for allele frequencies. Given a resource, and read evidence for the variant, Mutect2 will still emit variant alleles with AF less than or equal to the --max-population-af.
- Recapitulate any special options used in somatic calling in the panel of normals sample calling, e.g.
--disable-read-filter MateOnSameContigOrNoMappedMateReadFilter. This particular option is relevant for alt-aware and post-alt processed alignments.
Second, collate all the normal VCFs into a single callset with CreateSomaticPanelOfNormals. For the tutorial, to illustrate the step with small data, we run this command on three normal sample VCFs. The general recommendation for panel of normals is a minimum of forty samples.
gatk CreateSomaticPanelOfNormals \
-vcfs 3_HG00190.vcf.gz \
-vcfs 4_NA19771.vcf.gz \
-vcfs 5_HG02759.vcf.gz \
-O 6_threesamplepon.vcf.gz
This generates a PoN VCF 6_threesamplepon.vcf.gz and an index. The tutorial PoN contains 8,275 records.
CreateSomaticPanelOfNormals retains sites with variants in two or more samples. It retains the alleles from the samples but drops all other annotations to create an eight-column, sites-only VCF as shown.
Ideally, the PoN includes samples that are technically representative of the tumor case sample--i.e. samples sequenced on the same platform using the same chemistry, e.g. exome capture kit, and analyzed using the same toolchain. However, even an unmatched PoN will be remarkably effective in filtering a large proportion of sequencing artifacts. This is because mapping artifacts and polymerase slippage errors occur for pretty much the same genomic loci for short read sequencing approaches.
What do you think of including samples of family members in the PoN?
☞ 2.1 The tumor-only mode of Mutect2 is useful outside of pon creation
For example, consider variant calling on data that represents a pool of individuals or a collective of highly similar but distinct DNA molecules, e.g. mitochondrial DNA. Mutect2 calls multiple variants at a site in a computationally efficient manner. Furthermore, the tumor-only mode can be co-opted to simply call differences between two samples. This approach is described in Blog#11315.
back to top
3. Estimate cross-sample contamination using GetPileupSummaries and CalculateContamination.
First, run GetPileupSummaries on the tumor BAM to summarize read support for a set number of known variant sites. Use a population germline resource containing only common biallelic variants, e.g. subset by using SelectVariants --restrict-alleles-to BIALLELIC, as well as population AF allele frequencies in the INFO field [4]. The tool tabulates read counts that support reference, alternate and other alleles for the sites in the resource.
gatk GetPileupSummaries \
-I tumor.bam \
-V resources/chr17_small_exac_common_3_grch38.vcf.gz \
-O 7_tumor_getpileupsummaries.table
This produces a six-column table as shown. The alt_count is the count of reads that support the ALT allele in the germline resource. The allele_frequency corresponds to that given in the germline resource. Counts for other_alt_count refer to reads that support all other alleles.
Comments on select parameters
- The tool only considers homozygous alternate sites in the sample that have a population allele frequency that ranges between that set by
--minimum-population-allele-frequency (default 0.01) and --maximum-population-allele-frequency (default 0.2). The rationale for these settings is as follows. If the homozygous alternate site has a rare allele, we are more likely to observe the presence of REF or other more common alleles if there is cross-sample contamination. This allows us to measure contamination more accurately.
- One option to speed up analysis, that is not used in the command above, is to limit data collection to a sufficiently large but subset genomic region with the
-L argument.
Second, estimate contamination with CalculateContamination. The tool takes the summary table from GetPileupSummaries and gives the fraction contamination. This estimation informs downstream filtering by FilterMutectCalls.
gatk CalculateContamination \
-I 7_tumor_getpileupsummaries.table \
-O 8_tumor_calculatecontamination.table
This produces a table with estimates for contamination and error. The estimate for the full tumor sample is shown below and gives a contamination fraction of 0.0205. Going forward, we know to suspect calls with less than ~2% alternate allele fraction.
Comments on select parameters
- CalculateContamination can operate in two modes. The command above uses the mode that simply estimates contamination for a given sample. The alternate mode incorporates the metrics for the matched normal, to enable a potentially more accurate estimate. For the second mode, run GetPileupSummaries on the normal sample and then provide the normal pileup table to CalculateContamination with the
-matched argument.
► Cross-sample contamination differs from normal contamination of tumor and tumor contamination of normal. Currently, the workflow does not account for the latter type of purity issue.
☞ 3.1 What if I find high levels of contamination?
One thing to rule out is sample swaps at the read group level.
Picard’s CrosscheckFingerprints can detect sample-swaps at the read group level and can additionally measure how related two samples are. Because sequencing can involve multiplexing a sample across lanes and regrouping a sample’s multiple read groups, depending on the level of automation in handling these, there is a possibility of including read groups from unrelated samples. The inclusion of such a cross-sample in the tumor sample would be detrimental to a somatic analysis. Without getting into details, the tool allows us to (i) check at the sample level that our tumor and normal are related, as it is imperative they should come from the same individual and (ii) check at the read group level that each of the read group data come from the same individual.
Again, imagine if we mistook the contaminating read group data as some tumor subpopulation! The tutorial normal and tumor samples consist of 16 and 22 read groups respectively, and when we provide these and set EXPECT_ALL_GROUPS_TO_MATCH=true, CrosscheckReadGroupFingerprints (a tool now replaced by CrosscheckFingerprints) informs us All read groups related as expected.
back to top
4. Filter for confident somatic calls using FilterMutectCalls
FilterMutectCalls determines whether a call is a confident somatic call. The tool uses the annotations within the callset and applies preset thresholds that are tuned for human somatic analyses.
Filter the Mutect2 callset with FilterMutectCalls. Here we use the full callset, somatic_m2.vcf.gz. To activate filtering based on the contamination estimate, provide the contamination table with --contamination-table. In GATK v4.0.0.0, the tool uses the contamination estimate as a hard cutoff.
gatk FilterMutectCalls \
-V somatic_m2.vcf.gz \
--contamination-table tumor_calculatecontamination.table \
-O 9_somatic_oncefiltered.vcf.gz
This produces a VCF callset 9_somatic_oncefiltered.vcf.gz and index. Calls that are likely true positives get the PASS label in the FILTER field, and calls that are likely false positives are labeled with the reason(s) for filtering in the FILTER field of the VCF. We can view the available filters in the VCF header using grep '##FILTER'.
This step seemingly applies 14 filters, including contamination. However, if an annotation a filter relies on is absent, the tool skips the particular filtering. The filter will still appear in the header. For example, the duplicate_evidence filter requires a nonstandard annotation that our callset omits.
So far, we have 3,695 calls, of which 2,966 are filtered and 729 pass as confident somatic calls. Of the filtered, contamination filters eight calls, all of which would have been filtered for other reasons. For the statistically inclined, this may come as a surprise. However, remember that the great majority of contaminant variants would be common germline alleles, for which we have in place other safeguards.
► In the next GATK version, FilterMutectCalls will use a statistical model to filter based on the contamination estimate.
back to top
5. (Optional) Estimate artifacts with CollectSequencingArtifactMetrics and filter them with FilterByOrientationBias
FilterByOrientationBias allows filtering based on sequence context artifacts, e.g. OxoG and FFPE. This step is optional and if employed, should always be performed after filtering with FilterMutectCalls. The tool requires the pre_adapter_detail_metrics from Picard CollectSequencingArtifactMetrics.
First, collect metrics on sequence context artifacts with CollectSequencingArtifactMetrics. The tool categorizes these as those that occur before hybrid selection (preadapter) and those that occur during hybrid selection (baitbias). Results provide a global view across the genome that empowers decision making in ways that site-specific analyses cannot. The metrics can help decide whether to consider downstream filtering.
gatk CollectSequencingArtifactMetrics \
-I tumor.bam \
-O 10_tumor_artifact \
–-FILE_EXTENSION ".txt" \
-R ~/Documents/ref/hg38/Homo_sapiens_assembly38.fasta
Alternatively, use the tool from a standalone Picard jar.
java -jar picard.jar \
CollectSequencingArtifactMetrics \
I=tumor.bam \
O=10_tumor_artifact \
FILE_EXTENSION=.txt \
R=~/Documents/ref/hg38/Homo_sapiens_assembly38.fasta
This generates five metrics files, including pre_adapter_detail_metrics, which contains counts that FilterByOrientationBias uses. Below are the summary pre_adapter_summary_metrics for the full data. Our samples were not from FFPE so we do not expect this artifact. However, it appears that we could have some OxoG transversions.
![image]()
![image]()
Picard metrics are described in detail here. For the purposes of this tutorial, we focus on the TOTAL_QSCORE.
- The TOTAL_QSCORE is Phred-scaled such that lower scores equate to a higher probability the change is artifactual. E.g. forty translates to 1 in 10,000 probability. For OxoG, a rough cutoff for concern is 30. FilterByOrientationBias uses the quality score as a prior that a context will produce an artifact. The tool also weighs the evidence from the reads. For example, if the QSCORE is 50 but the allele is supported by 15 reads in F1R2 and no reads in F2R1, then the tool should filter the call.
- FFPE stands for formalin-fixed, paraffin-embedded. Formaldehyde deaminates cytosines and thereby results in C→T transition mutations. Oxidation of guanine to 8-oxoguanine results in G→T transversion mutations during library preparation. Another Picard tool, CollectOxoGMetrics, similarly gives Phred-scaled scores for the 16 three-base extended sequence contexts. In GATK4 Mutect2, the
F1R2 and F2R1 annotations count the reads in the pair orientation supporting the allele(s). This is a change from GATK3’s FOXOG (fraction OxoG) annotation.
Second, perform orientation bias filtering with FilterByOrientationBias. We provide the tool with the once-filtered calls 9_somatic_oncefiltered.vcf.gz, the pre_adapter_detail_metrics file and the sequencing contexts for FFPE (C→T transition) and OxoG (G→T transversion). The tool knows to include the reverse complement contexts.
gatk FilterByOrientationBias \
-A G/T \
-A C/T \
-V 9_somatic_oncefiltered.vcf.gz \
-P tumor_artifact.pre_adapter_detail_metrics.txt \
-O 11_somatic_twicefiltered.vcf.gz
This produces a VCF 11_somatic_twicefiltered.vcf.gz, index and summary 11_somatic_twicefiltered.vcf.gz.summary. In the summary, we see the number of calls for the sequence context and the number of those that the tool filters.
Is the filtering in line with our earlier prediction?
In the VCF header, we see the addition of the 15th filter, orientation_bias, which the tool applies to 56 calls. All 56 of these calls were previously PASS sites, i.e. unfiltered. We now have 673 passing calls out of 3,695 total calls.
☞ 5.1 Tally of applied filters for the tutorial data
The table shows the breakdown in filters applied to 11_somatic_twicefiltered.vcf.gz. The middle column tallys the instances in which each filter was applied across the calls and the third column tallys the instances in which a filter was the sole reason for a site not passing. Of the total calls, ~18% (673/3,695) are confident somatic calls. Of the filtered calls, ~56% (1,694/3,022) are filtered singly. We see an average of ~1.73 filters per filtered call (5,223/3,022).
Which filters appear to have the greatest impact? What types of calls do you think compels manual review?
Examine passing records with the following command. Take note of the AD and AF annotation values in particular, as they show the high sensitivity of the caller.
gzcat 11_somatic_twicefiltered.vcf.gz | grep -v '#' | awk '$7=="PASS"' | less
back to top
6. Set up in IGV to review somatic calls
Deriving a good somatic callset involves comparing callsets, e.g. from different callers or calling approaches, manually reviewing passing and filtered calls and, if necessary, combining callsets and additional filtering. Manual review extends from deciphering call record annotations to the nitty-gritty of reviewing read alignments using a visualizer.
To manually review calls, use the feature-rich desktop version of the Integrative Genomics Viewer (IGV). Remember that Mutect2 makes calls on reassembled alignments that do not necessarily reflect that of the starting BAM. Given this, viewing the raw BAM is insufficient for understanding calls. We must examine the bamout that Mutect2's graph-assembly produces.
First, load Human (hg38) as the reference in IGV. Then load these six files in order:
- resources/chr17_pon.vcf.gz
- resources/chr17_af-only-gnomad_grch38.vcf.gz
- 11_somatic_twicefiltered.vcf.gz
- 2_tumor_normal_m2.bam
- normal.bam
- tumor.bam
With the exception of the somatic callset 11_somatic_twicefiltered.vcf.gz, the subset regions the data cover are in chr17plus.interval_list.
![image]() Second, navigate IGV to the TP53 locus (chr17:7,666,402-7,689,550).
Second, navigate IGV to the TP53 locus (chr17:7,666,402-7,689,550).
- One of the tracks is dominating the view. Right-click on track
chr17_af-only-gnomad_grch38.vcf.gz and collapse its view.
![image]() Zoom into the somatic call in
Zoom into the somatic call in 11_somatic_twicefiltered.vcf.gz, the gray rectangle in exon 3, by click-dragging on the ruler.- Hover over or click on the gray call in track
11_somatic_twicefiltered.vcf.gz to view INFO level annotations. Similarly, the blue call underneath gives HCC1143_tumor sample level information.
- Scroll through the alignment data and notice the coverage for the samples.
A C→T variant is in tumor.bam but not normal.bam. What is happening in 2_tumor_normal_m2.bam?
![image]() Third, tweak IGV settings that aid in visualizing reassembled alignments.
Third, tweak IGV settings that aid in visualizing reassembled alignments.
What are the three grouped tracks for the bamout? What does the pastel versus gray colors indicate? How plausible is it that all tumor copies of this locus have this alteration?
Here is the corresponding VCF record. Remember Mutect2 makes no ploidy assumption. The GT field tabulates the presence for each allele starting with the reference allele.
| CHROM |
POS |
ID |
REF |
ALT |
QUAL |
FILTER |
INFO |
| chr17 |
7,674,220 |
. |
C |
T |
. |
PASS |
DP=122;ECNT=1;NLOD=13.54;N_ART_LOD=-1.675e+00;POP_AF=2.500e-06;P_GERMLINE=-1.284e+01;TLOD=257.15 |
| FORMAT |
GT:AD:AF:F1R2:F2R1:MBQ:MFRL:MMQ:MPOS:OBAM:OBAMRC:OBF:OBP:OBQ:OBQRC:SA_MAP_AF:SA_POST_PROB |
| HCC1143_normal |
0/0:45,0:0.032:19,0:26,0:0:151,0:0:0:false:false |
| HCC1143_tumor |
0/1:0,70:0.973:0,34:0,36:33:0,147:60:21:true:false:0.486:0.00:46.01:100.00:0.990,0.990,1.00:0.028,0.026,0.946 |
Finally, here are the indel calls for which we have bamout alignments. All 17 of these happen to be filtered. Explore a few of these sites in IGV to practice the motions of setting up for manual review and to study the logic behind different filters.
| CHROM |
POS |
REF |
ALT |
FILTER |
| chr17 |
4,539,344 |
T |
TA |
artifact_in_normal;germline_risk;panel_of_normals |
| chr17 |
7,221,420 |
CACTGCCCTAGGTCAGGA |
C |
artifact_in_normal;panel_of_normals;str_contraction |
| chr17 |
7,483,063 |
A |
AC |
mapping_quality;t_lod |
| chr17 |
8,513,688 |
GTT |
G |
panel_of_normals |
| chr17 |
19,748,387 |
G |
GA |
t_lod |
| chr17 |
26,982,033 |
G |
GC |
artifact_in_normal;clustered_events |
| chr17 |
30,059,463 |
CT |
C |
t_lod |
| chr17 |
35,422,473 |
C |
CA |
t_lod |
| chr17 |
35,671,734 |
CTT |
C,CT,CTTT |
artifact_in_normal;multiallelic;panel_of_normals |
| chr17 |
43,104,057 |
CA |
C |
artifact_in_normal;germline_risk;panel_of_normals |
| chr17 |
43,104,072 |
AAAAAAAAAGAAAAG |
A |
panel_of_normals;t_lod |
| chr17 |
46,332,538 |
G |
GT |
artifact_in_normal;panel_of_normals |
| chr17 |
47,157,394 |
CAA |
C |
panel_of_normals;t_lod |
| chr17 |
50,124,771 |
GCACACACACACACACA |
G |
clustered_events;panel_of_normals;t_lod |
| chr17 |
68,907,890 |
GA |
G |
artifact_in_normal;base_quality;germline_risk;panel_of_normals;t_lod |
| chr17 |
69,182,632 |
C |
CA |
artifact_in_normal;t_lod |
| chr17 |
69,182,835 |
GAAAA |
G |
panel_of_normals |
back to top
7. Related resources
The next step after generating a carefully manicured somatic callset is typically functional annotation.
- Funcotator is available in BETA and can annotate GRCh38 and prior reference aligned VCF format data.
- Oncotator can annotate GRCh37 and prior reference aligned MAF and VCF format data. It is also possible to download and install the tool following instructions in Article#4154.
- Annotate with the external program VEP to predict phenotypic changes and confirm or hypothesize biochemical effects.
For a cohort, after annotation, use MutSig to discover driver mutations. MutsigCV (the version is CV) is available on GenePattern. If more samples are needed to increase the power of the analysis, consider padding the analysis set with TCGA Project or other data.
The dSKY plot at https://figshare.com/articles/D_SKY_for_HCC1143/2056665 shows somatic copy number alterations for the HCC1143 tumor sample. Its colorful results remind us that calling SNVs and indels is only one part of cancer genome analyses. Somatic copy number alteration detection will be covered in another GATK tutorial. For reference implementations of Somatic CNV workflows see here.
back to top
Footnotes
[1] Data was alt-aware aligned to GRCh38 and post-alt processed. For an introduction to alt-aware alignment and post-alt processing, see [Blog#8180](https://software.broadinstitute.org/gatk/blog?id=8180). The HCC1143 alignments are identical to that in [Tutorial#9183](https://software.broadinstitute.org/gatk/documentation/article?id=9183), which uses GATK3 MuTect2.
[2] For scripted GATK Best Practices Somatic Short Variant Discovery workflows, see [https://github.com/gatk-workflows](https://github.com/gatk-workflows). Within the repository, as of this writing, [gatk-somatic-snvs-indels](https://github.com/gatk-workflows/gatk4-somatic-snvs-indels), which uses GRCh37, is the sole GATK4 Mutect2 workflow. This tutorial uses additional parameters not used in the [GRCh37 gatk-somatic-snvs-indels](https://github.com/gatk-workflows/gatk4-somatic-snvs-indels) example because the tutorial data was preprocessed with post-alt processing of alt-aware alignments, which deviates from production practices. The general workflow steps remain the same.
[3] About the tutorial data:
- The data tarball contains 15 files in the main directory, six files in its resources folder and twenty files in its precomputed folder. Of the files, chr17 refers to data subset to that in the regions in
chr17plus.interval_list, the m2pon consists of forty 1000 Genomes Project samples, pon to panel of normals, tumor to the tumor HCC1143 breast cancer sample and normal to its matched blood normal.
- Again, example data are based on a breast cancer cell line and its matched normal cell line derived from blood. Both cell lines are consented and known as HCC1143 and HCC1143_BL, respectively. The Broad Cancer Genome Analysis (CGA) group has graciously provided 2x76 paired-end whole exome sequence data from the two cell lines (C835.HCC1143_2 and C835.HCC1143_BL.4), and @shlee reverted and aligned these to GRCh38 using alt-aware alignment and post-alt processing as described in Tutorial#8017. During preprocessing, the MergeBamAlignment step was omitted, reads containing adapter sequence were removed altogether for both samples (~0.153% of reads in the tumor) as determined by MarkIlluminaAdapters, base qualities were not binned during base recalibration and indel realignment was included to match the toolchain of the PoN normals. The program group for base recalibration is absent from the BAM headers due to a bug in the version of PrintReads at the time of pre-processing, in January of 2017.
- Note that the tutorial uses exome data for its small size. The workflow is applicable to whole genome sequence data (WGS).
- @shlee lifted-over or remapped the gnomAD resource files from GRCh37 counterparts to GRCh38. The tutorial uses subsets of the full resources; the full-length versions are available at gs://gatk-best-practices/somatic-hg38/. The official GRCh37 versions of the resources are available in the GATK Resource Bundle and are based on the gnomAD resource. These GRCh37 versions were prepared by @davidben according to the method outlined in the mutect_resources.wdl and described in [4].
- The full data in the tutorial were generated by @shlee using the github.com/broadinstitute/gatk mutect2.wdl from between the v4.0.0.0 and v4.0.0.1 release with commit hash
b4d1ddd. The GATK Docker image was broadinstitute/gatk:4.0.0.0 and Picard was v2.14.1. A single modification was made to the script to enable generating the bamout. The script was run locally on a Google Cloud Compute VM using Cromwell v30.1. Given Docker was installed and the specified Docker images were present on the VM, Cromwell automatically launched local Docker container instances during the run and handled the local files as hard-links to avoid redundant copying. Workflow input variables were as follows.
{
"##_COMMENT1:": "WORKFLOW STEP OPTIONS",
"Mutect2.is_run_oncotator": "False",
"Mutect2.is_run_orientation_bias_filter": "True",
"Mutect2.picard": "/home/shlee/picard-2.14.1.jar",
"Mutect2.gatk_docker": "broadinstitute/gatk:4.0.0.0",
"Mutect2.oncotator_docker": "broadinstitute/oncotator:1.9.3.0",
...
"##_COMMENT3:": "ANALYSIS PARAMETERS",
"Mutect2.artifact_modes": ["G/T", "C/T"],
"Mutect2.m2_extra_args": "--af-of-alleles-not-in-resource 0.0000025 --disable-read-filter MateOnSameContigOrNoMappedMateReadFilter",
"Mutect2.m2_extra_filtering_args": "",
"Mutect2.scatter_count": "10"
}
- If using newer versions of the
mutect2.wdl that allow setting SplitIntervals optional arguments, then @shlee recommends setting --subdivision-mode BALANCING_WITHOUT_INTERVAL_SUBDIVISION to avoid splitting contigs.
- With the exception of the PoN and Picard tool steps, data was generated using v4.0.0.0. The PoN was generated using GATK4 vbeta.6. Besides the syntax, little changed for the Mutect2 workflow between these releases and the workflow and most of its tools remain in beta status as of this writing. We used Picard v2.14.1 for the CollectSequencingArtifactMetrics step. Figures in section 5 reflect results from Picard v2.11.0, which give, at glance, identical results as 2.14.1.
- The three samples in section 2 are present in the forty sample PoN used in section 1 and they are 1000 Genomes Project samples.
[4] The WDL script [mutect_resources.wdl](https://github.com/broadinstitute/gatk/blob/master/scripts/mutect2_wdl/mutect_resources.wdl) takes a large gnomAD VCF or other typical cohort VCF and from it prepares both a simplified germline resource for use in _section 1_ and a common biallelic variants resource for use in _section 3_. The script first generates a sites-only VCF and in the process _removes all extraneous annotations_ except for `AF` allele frequencies. We recommend this simplification as the unburdened VCF allows Mutect2 to run much more efficiently. To generate the common biallelic variants resource, the script then selects the biallelic sites from the sites-only VCF.
back to top

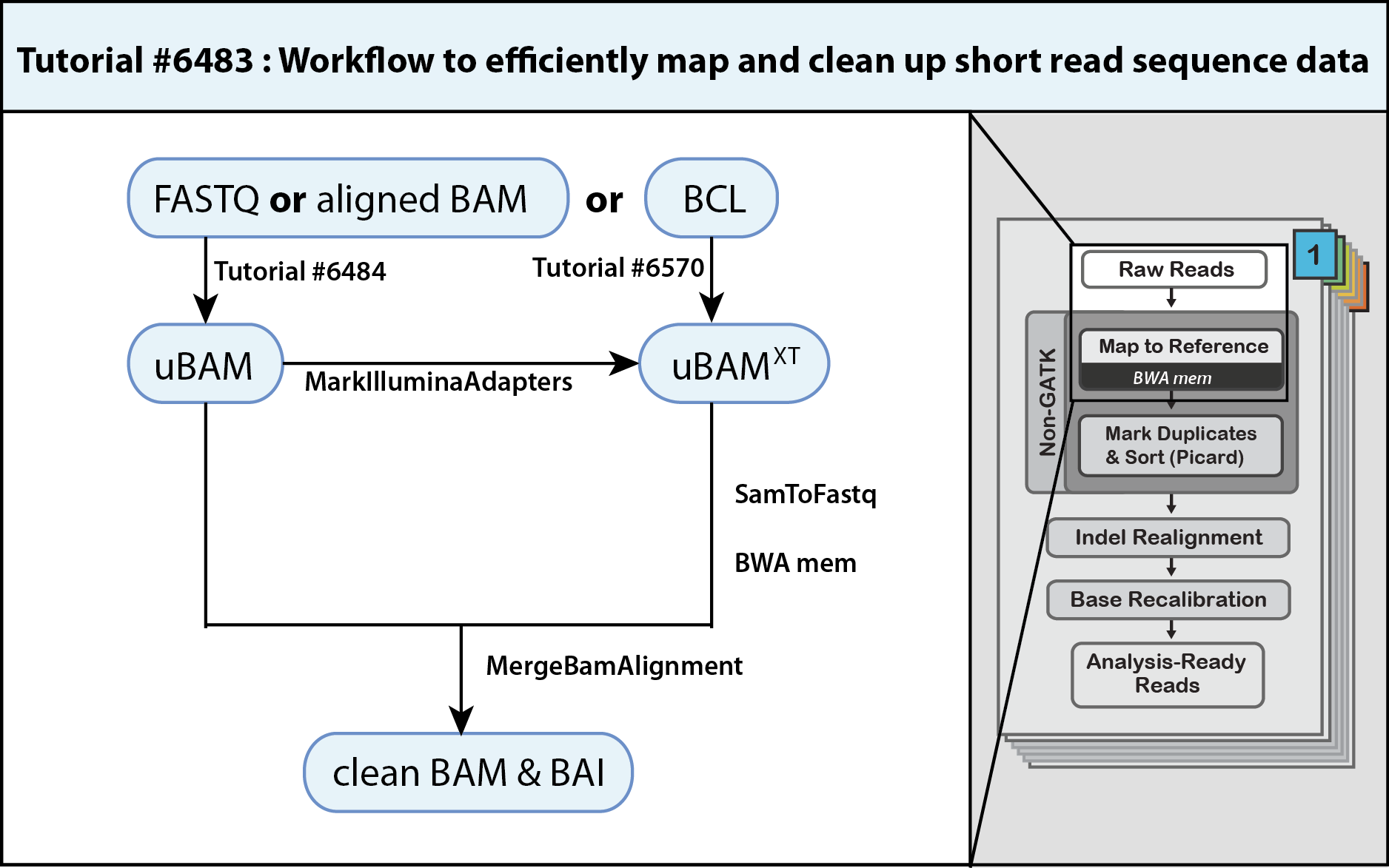 If you are interested in emulating the methods used by the Broad Genomics Platform to pre-process your short read sequencing data, you have landed on the right page. The parsimonious operating procedures outlined in this three-step workflow both maximize data quality, storage and processing efficiency to produce a mapped and clean BAM. This clean BAM is ready for analysis workflows that start with MarkDuplicates.
If you are interested in emulating the methods used by the Broad Genomics Platform to pre-process your short read sequencing data, you have landed on the right page. The parsimonious operating procedures outlined in this three-step workflow both maximize data quality, storage and processing efficiency to produce a mapped and clean BAM. This clean BAM is ready for analysis workflows that start with MarkDuplicates.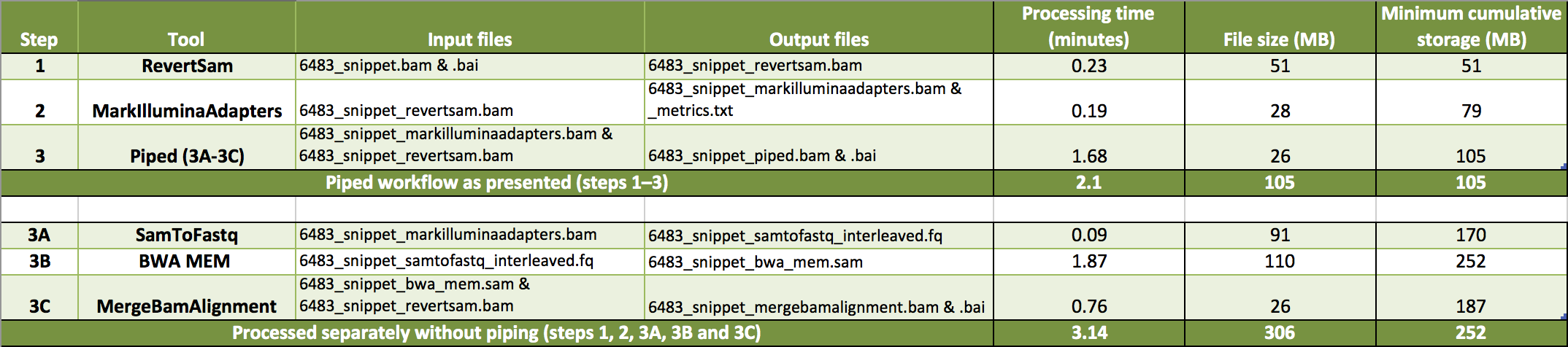









 A read with multiple alignment records may map to multiple loci or may be chimeric--that is, splits the alignment. It is possible for an aligner to produce multiple alignments as well as multiple primary alignments, e.g. in the case of a linear alignment set of split reads. When one alignment, or alignment set in the case of chimeric read records, is designated primary, others are designated either secondary or supplementary. Invoking the
A read with multiple alignment records may map to multiple loci or may be chimeric--that is, splits the alignment. It is possible for an aligner to produce multiple alignments as well as multiple primary alignments, e.g. in the case of a linear alignment set of split reads. When one alignment, or alignment set in the case of chimeric read records, is designated primary, others are designated either secondary or supplementary. Invoking the  Tags are updated for our example data as shown in the table. The tool retains SA, MD, NM and AS tags from the alignment, given these are not present in the uBAM. The tool additionally adds UQ (the Phred likelihood of the segment), MC (mate CIGAR string) and MQ (mapping quality of the mate/next segment) tags if applicable. For unmapped reads (marked with an
Tags are updated for our example data as shown in the table. The tool retains SA, MD, NM and AS tags from the alignment, given these are not present in the uBAM. The tool additionally adds UQ (the Phred likelihood of the segment), MC (mate CIGAR string) and MQ (mapping quality of the mate/next segment) tags if applicable. For unmapped reads (marked with an 

 We pipe the three tools described above to generate an aligned BAM file sorted by query name. In the piped command, the commands for the three processes are given together, separated by a
We pipe the three tools described above to generate an aligned BAM file sorted by query name. In the piped command, the commands for the three processes are given together, separated by a 

















