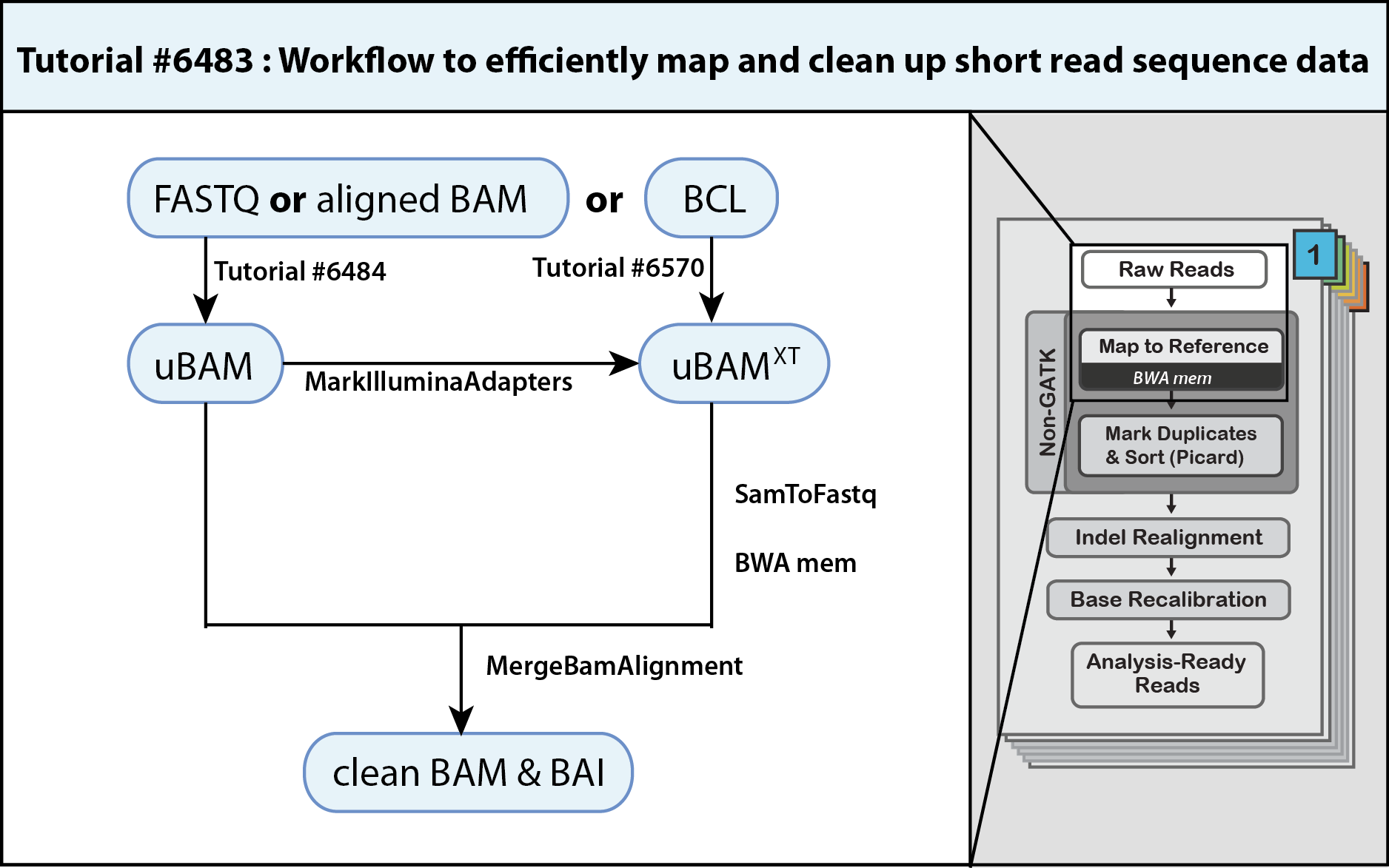
![image]() If you are interested in emulating the methods used by the Broad Genomics Platform to pre-process your short read sequencing data, you have landed on the right page. The parsimonious operating procedures outlined in this three-step workflow both maximize data quality, storage and processing efficiency to produce a mapped and clean BAM. This clean BAM is ready for analysis workflows that start with MarkDuplicates.
If you are interested in emulating the methods used by the Broad Genomics Platform to pre-process your short read sequencing data, you have landed on the right page. The parsimonious operating procedures outlined in this three-step workflow both maximize data quality, storage and processing efficiency to produce a mapped and clean BAM. This clean BAM is ready for analysis workflows that start with MarkDuplicates.
Since your sequencing data could be in a number of formats, the first step of this workflow refers you to specific methods to generate a compatible unmapped BAM (uBAM, Tutorial#6484) or (uBAMXT, Tutorial#6570 coming soon). Not all unmapped BAMs are equal and these methods emphasize cleaning up prior meta information while giving you the opportunity to assign proper read group fields. The second step of the workflow has you marking adapter sequences, e.g. arising from read-through of short inserts, using MarkIlluminaAdapters such that they contribute minimally to alignments and allow the aligner to map otherwise unmappable reads. The third step pipes three processes to produce the final BAM. Piping SamToFastq, BWA-MEM and MergeBamAlignment saves time and allows you to bypass storage of larger intermediate FASTQ and SAM files. In particular, MergeBamAlignment merges defined information from the aligned SAM with that of the uBAM to conserve read data, and importantly, it generates additional meta information and unifies meta data. The resulting clean BAM is coordinate sorted, indexed.
The workflow reflects a lossless operating procedure that retains original sequencing read information within the final BAM file such that data is amenable to reversion and analysis by different means. These practices make scaling up and long-term storage efficient, as one needs only keep the final BAM file.
Geraldine_VdAuwera points out that there are many different ways of correctly preprocessing HTS data for variant discovery and ours is only one approach. So keep this in mind.
We present this workflow using real data from a public sample. The original data file, called Solexa-272222, is large at 150 GB. The file contains 151 bp paired PCR-free reads giving 30x coverage of a human whole genome sample referred to as NA12878. The entire sample library was sequenced in a single flow cell lane and thereby assigns all the reads the same read group ID. The example commands work both on this large file and on smaller files containing a subset of the reads, collectively referred to as snippet. NA12878 has a variant in exon 5 of the CYP2C19 gene, on the portion of chromosome 10 covered by the snippet, resulting in a nonfunctional protein. Consistent with GATK's recommendation of using the most up-to-date tools, for the given example results, with the exception of BWA, we used the most current versions of tools as of their testing (September to December 2015). We provide illustrative example results, some of which were derived from processing the original large file and some of which show intermediate stages skipped by this workflow.
Download example snippet data to follow along the tutorial.
We welcome feedback. Share your suggestions in the Comments section at the bottom of this page.
Jump to a section
- Generate an unmapped BAM from FASTQ, aligned BAM or BCL
- Mark adapter sequences using MarkIlluminaAdapters
- Align reads with BWA-MEM and merge with uBAM using MergeBamAlignment
A. Convert BAM to FASTQ and discount adapter sequences using SamToFastq
B. Align reads and flag secondary hits using BWA-MEM
C. Restore altered data and apply & adjust meta information using MergeBamAlignment
D. Pipe SamToFastq, BWA-MEM and MergeBamAlignment to generate a clean BAM
Tools involved
Prerequisites
- Installed Picard tools
- Installed GATK tools
- Installed BWA
- Reference genome
- Illumina or similar tech DNA sequence reads file containing data corresponding to one read group ID. That is, the file contains data from one sample and from one flow cell lane.
Download example data
- To download the reference, open ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/2.8/b37/ in your browser. Leave the password field blank. Download the following three files (~860 MB) to the same folder:
human_g1k_v37_decoy.fasta.gz, .fasta.fai.gz, and .dict.gz. This same reference is available to load in IGV.
- I divided the example data into two tarballs: tutorial_6483_piped.tar.gz contains the files for the piped process and tutorial_6483_intermediate_files.tar.gz contains the intermediate files produced by running each process independently. The data contain reads originally aligning to a one Mbp genomic interval (10:96,000,000-97,000,000) of GRCh37. The table shows the steps of the workflow, corresponding input and output example data files and approximate minutes and disk space needed to process each step. Additionally, we tabulate the time and minimum storage needed to complete the workflow as presented (piped) or without piping.
![image]()
Related resources
Other notes
- When transforming data files, we stick to using Picard tools over other tools to avoid subtle incompatibilities.
For large files, (1) use the Java -Xmx setting and (2) set the environmental variable TMP_DIR for a temporary directory.
java -Xmx8G -jar /path/picard.jar MarkIlluminaAdapters \
TMP_DIR=/path/shlee
In the command, the -Xmx8G Java option caps the maximum heap size, or memory usage, to eight gigabytes. The path given by TMP_DIR points the tool to scratch space that it can use. These options allow the tool to run without slowing down as well as run without causing an out of memory error. The -Xmx settings we provide here are more than sufficient for most cases. For GATK, 4G is standard, while for Picard less is needed. Some tools, e.g. MarkDuplicates, may require more. These options can be omitted for small files such as the example data and the equivalent command is as follows.
java -jar /path/picard.jar MarkIlluminaAdapters
To find a system's default maximum heap size, type java -XX:+PrintFlagsFinal -version, and look for MaxHeapSize. Note that any setting beyond available memory spills to storage and slows a system down. If multithreading, increase memory proportionately to the number of threads. e.g. if 1G is the minimum required for one thread, then use 2G for two threads.
When I call default options within a command, follow suit to ensure the same results.
1. Generate an unmapped BAM from FASTQ, aligned BAM or BCL
If you have raw reads data in BAM format with appropriately assigned read group fields, then you can start with step 2. Namely, besides differentiating samples, the read group ID should differentiate factors contributing to technical batch effects, i.e. flow cell lane. If not, you need to reassign read group fields. This dictionary post describes factors to consider and this post and this post provide some strategic advice on handling multiplexed data.
If your reads are mapped, or in BCL or FASTQ format, then generate an unmapped BAM according to the following instructions.
- To convert FASTQ or revert aligned BAM files, follow directions in Tutorial#6484. The resulting uBAM needs to have its adapter sequences marked as outlined in the next step (step 2).
- To convert an Illumina Base Call files (BCL) use IlluminaBasecallsToSam. The tool marks adapter sequences at the same time. The resulting uBAMXT has adapter sequences marked with the XT tag so you can skip step 2 of this workflow and go directly to step 3. The corresponding Tutorial#6570 is coming soon.
See if you can revert 6483_snippet.bam, containing 279,534 aligned reads, to the unmapped 6383_snippet_revertsam.bam, containing 275,546 reads.
back to top
2. Mark adapter sequences using MarkIlluminaAdapters
MarkIlluminaAdapters adds the XT tag to a read record to mark the 5' start position of the specified adapter sequence and produces a metrics file. Some of the marked adapters come from concatenated adapters that randomly arise from the primordial soup that is a PCR reaction. Others represent read-through to 3' adapter ends of reads and arise from insert sizes that are shorter than the read length. In some instances read-though can affect the majority of reads in a sample, e.g. in Nextera library samples over-titrated with transposomes, and render these reads unmappable by certain aligners. Tools such as SamToFastq use the XT tag in various ways to effectively remove adapter sequence contribution to read alignment and alignment scoring metrics. Depending on your library preparation, insert size distribution and read length, expect varying amounts of such marked reads.
java -Xmx8G -jar /path/picard.jar MarkIlluminaAdapters \
I=6483_snippet_revertsam.bam \
O=6483_snippet_markilluminaadapters.bam \
M=6483_snippet_markilluminaadapters_metrics.txt \ #naming required
TMP_DIR=/path/shlee #optional to process large files
This produces two files. (1) The metrics file, 6483_snippet_markilluminaadapters_metrics.txt bins the number of tagged adapter bases versus the number of reads. (2) The 6483_snippet_markilluminaadapters.bam file is identical to the input BAM, 6483_snippet_revertsam.bam, except reads with adapter sequences will be marked with a tag in XT:i:# format, where # denotes the 5' starting position of the adapter sequence. At least six bases are required to mark a sequence. Reads without adapter sequence remain untagged.
- By default, the tool uses Illumina adapter sequences. This is sufficient for our example data.
- Adjust the default standard Illumina adapter sequences to any adapter sequence using the
FIVE_PRIME_ADAPTER and THREE_PRIME_ADAPTER parameters. To clear and add new adapter sequences first set ADAPTERS to 'null' then specify each sequence with the parameter.
We plot the metrics data that is in GATKReport file format using RStudio, and as you can see, marked bases vary in size up to the full length of reads.
![image]()
![image]()
Do you get the same number of marked reads? 6483_snippet marks 448 reads (0.16%) with XT, while the original Solexa-272222 marks 3,236,552 reads (0.39%).
Below, we show a read pair marked with the XT tag by MarkIlluminaAdapters. The insert region sequences for the reads overlap by a length corresponding approximately to the XT tag value. For XT:i:20, the majority of the read is adapter sequence. The same read pair is shown after SamToFastq transformation, where adapter sequence base quality scores have been set to 2 (# symbol), and after MergeBamAlignment, which restores original base quality scores.
Unmapped uBAM (step 1)
![image]()
After MarkIlluminaAdapters (step 2)
![image]()
After SamToFastq (step 3)
![image]()
After MergeBamAlignment (step 3)
![image]()
back to top
3. Align reads with BWA-MEM and merge with uBAM using MergeBamAlignment
This step actually pipes three processes, performed by three different tools. Our tutorial example files are small enough to easily view, manipulate and store, so any difference in piped or independent processing will be negligible. For larger data, however, using Unix pipelines can add up to significant savings in processing time and storage.
Not all tools are amenable to piping and piping the wrong tools or wrong format can result in anomalous data.
The three tools we pipe are SamToFastq, BWA-MEM and MergeBamAlignment. By piping these we bypass storage of larger intermediate FASTQ and SAM files. We additionally save time by eliminating the need for the processor to read in and write out data for two of the processes, as piping retains data in the processor's input-output (I/O) device for the next process.
To make the information more digestible, we will first talk about each tool separately. At the end of the section, we provide the piped command.
back to top
3A. Convert BAM to FASTQ and discount adapter sequences using SamToFastq
Picard's SamToFastq takes read identifiers, read sequences, and base quality scores to write a Sanger FASTQ format file. We use additional options to effectively remove previously marked adapter sequences, in this example marked with an XT tag. By specifying CLIPPING_ATTRIBUTE=XT and CLIPPING_ACTION=2, SamToFastq changes the quality scores of bases marked by XT to two--a rather low score in the Phred scale. This effectively removes the adapter portion of sequences from contributing to downstream read alignment and alignment scoring metrics.
Illustration of an intermediate step unused in workflow. See piped command.
java -Xmx8G -jar /path/picard.jar SamToFastq \
I=6483_snippet_markilluminaadapters.bam \
FASTQ=6483_snippet_samtofastq_interleaved.fq \
CLIPPING_ATTRIBUTE=XT \
CLIPPING_ACTION=2 \
INTERLEAVE=true \
NON_PF=true \
TMP_DIR=/path/shlee #optional to process large files
This produces a FASTQ file in which all extant meta data, i.e. read group information, alignment information, flags and tags are purged. What remains are the read query names prefaced with the @ symbol, read sequences and read base quality scores.
- For our paired reads example file we set SamToFastq's
INTERLEAVE to true. During the conversion to FASTQ format, the query name of the reads in a pair are marked with /1 or /2 and paired reads are retained in the same FASTQ file. BWA aligner accepts interleaved FASTQ files given the -p option.
- We change the
NON_PF, aka INCLUDE_NON_PF_READS, option from default to true. SamToFastq will then retain reads marked by what some consider an archaic 0x200 flag bit that denotes reads that do not pass quality controls, aka reads failing platform or vendor quality checks. Our tutorial data do not contain such reads and we call out this option for illustration only.
- Other CLIPPING_ACTION options include (1) X to hard-clip, (2) N to change bases to Ns or (3) another number to change the base qualities of those positions to the given value.
back to top
3B. Align reads and flag secondary hits using BWA-MEM
In this workflow, alignment is the most compute intensive and will take the longest time. GATK's variant discovery workflow recommends Burrows-Wheeler Aligner's maximal exact matches (BWA-MEM) algorithm (Li 2013 reference; Li 2014 benchmarks; homepage; manual). BWA-MEM is suitable for aligning high-quality long reads ranging from 70 bp to 1 Mbp against a large reference genome such as the human genome.
- Aligning our
snippet reads against either a portion or the whole genome is not equivalent to aligning our original Solexa-272222 file, merging and taking a new slice from the same genomic interval.
- For the tutorial, we use BWA v 0.7.7.r441, the same aligner used by the Broad Genomics Platform as of this writing (9/2015).
- As mentioned, alignment is a compute intensive process. For faster processing, use a reference genome with decoy sequences, also called a decoy genome. For example, the Broad's Genomics Platform uses an Hg19/GRCh37 reference sequence that includes Ebstein-Barr virus (EBV) sequence to soak up reads that fail to align to the human reference that the aligner would otherwise spend an inordinate amount of time trying to align as split reads. GATK's resource bundle provides a standard decoy genome from the 1000 Genomes Project.
BWA alignment requires an indexed reference genome file. Indexing is specific to algorithms. To index the human genome for BWA, we apply BWA's index function on the reference genome file, e.g. human_g1k_v37_decoy.fasta. This produces five index files with the extensions amb, ann, bwt, pac and sa.
bwa index -a bwtsw human_g1k_v37_decoy.fasta
The example command below aligns our example data against the GRCh37 genome. The tool automatically locates the index files within the same folder as the reference FASTA file.
Illustration of an intermediate step unused in workflow. See piped command.
/path/bwa mem -M -t 7 -p /path/human_g1k_v37_decoy.fasta \
6483_snippet_samtofastq_interleaved.fq > 6483_snippet_bwa_mem.sam
This command takes the FASTQ file, 6483_snippet_samtofastq_interleaved.fq, and produces an aligned SAM format file, 6483_snippet_unthreaded_bwa_mem.sam, containing read alignment information, an automatically generated program group record and reads sorted in the same order as the input FASTQ file. Aligner-assigned alignment information, flag and tag values reflect each read's or split read segment's best sequence match and does not take into consideration whether pairs are mapped optimally or if a mate is unmapped. Added tags include the aligner-specific XS tag that marks secondary alignment scores in XS:i:# format. This tag is given for each read even when the score is zero and even for unmapped reads. The program group record (@PG) in the header gives the program group ID, group name, group version and recapitulates the given command. Reads are sorted by query name. For the given version of BWA, the aligned file is in SAM format even if given a BAM extension.
Does the aligned file contain read group information?
We invoke three options in the command.
-M to flag shorter split hits as secondary.
This is optional for Picard compatibility as MarkDuplicates can directly process BWA's alignment, whether or not the alignment marks secondary hits. However, if we want MergeBamAlignment to reassign proper pair alignments, to generate data comparable to that produced by the Broad Genomics Platform, then we must mark secondary alignments.
-p to indicate the given file contains interleaved paired reads.
-t followed by a number for the number of processor threads to use concurrently. Here we use seven threads which is one less than the total threads available on my Mac laptap. Check your server or system's total number of threads with the following command provided by KateN.
getconf _NPROCESSORS_ONLN
In the example data, all of the 1211 unmapped reads each have an asterisk (*) in column 6 of the SAM record, where a read typically records its CIGAR string. The asterisk represents that the CIGAR string is unavailable. The several asterisked reads I examined are recorded as mapping exactly to the same location as their _mapped_ mates but with MAPQ of zero. Additionally, the asterisked reads had varying noticeable amounts of low base qualities, e.g. strings of #s, that corresponded to original base quality calls and not those changed by SamToFastq. This accounting by BWA allows these pairs to always list together, even when the reads are coordinate-sorted, and leaves a pointer to the genomic mapping of the mate of the unmapped read. For the example read pair shown below, comparing sequences shows no apparent overlap, with the highest identity at 72% over 25 nts.
After MarkIlluminaAdapters (step 2)
![image]()
After BWA-MEM (step 3)
![image]()
After MergeBamAlignment (step 3)
![image]()
back to top
3C. Restore altered data and apply & adjust meta information using MergeBamAlignment
MergeBamAlignment is a beast of a tool, so its introduction is longer. It does more than is implied by its name. Explaining these features requires I fill you in on some background.
Broadly, the tool merges defined information from the unmapped BAM (uBAM, step 1) with that of the aligned BAM (step 3) to conserve read data, e.g. original read information and base quality scores. The tool also generates additional meta information based on the information generated by the aligner, which may alter aligner-generated designations, e.g. mate information and secondary alignment flags. The tool then makes adjustments so that all meta information is congruent, e.g. read and mate strand information based on proper mate designations. We ascribe the resulting BAM as clean.
Specifically, the aligned BAM generated in step 3 lacks read group information and certain tags--the UQ (Phred likelihood of the segment), MC (CIGAR string for mate) and MQ (mapping quality of mate) tags. It has hard-clipped sequences from split reads and altered base qualities. The reads also have what some call mapping artifacts but what are really just features we should not expect from our aligner. For example, the meta information so far does not consider whether pairs are optimally mapped and whether a mate is unmapped (in reality or for accounting purposes). Depending on these assignments, MergeBamAlignment adjusts the read and read mate strand orientations for reads in a proper pair. Finally, the alignment records are sorted by query name. We would like to fix all of these issues before taking our data to a variant discovery workflow.
Enter MergeBamAlignment. As the tool name implies, MergeBamAlignment applies read group information from the uBAM and retains the program group information from the aligned BAM. In restoring original sequences, the tool adjusts CIGAR strings from hard-clipped to soft-clipped. If the alignment file is missing reads present in the unaligned file, then these are retained as unmapped records. Additionally, MergeBamAlignment evaluates primary alignment designations according to a user-specified strategy, e.g. for optimal mate pair mapping, and changes secondary alignment and mate unmapped flags based on its calculations. Additional for desired congruency. I will soon explain these and additional changes in more detail and show a read record to illustrate.
Consider what PRIMARY_ALIGNMENT_STRATEGY option best suits your samples. MergeBamAlignment applies this strategy to a read for which the aligner has provided more than one primary alignment, and for which one is designated primary by virtue of another record being marked secondary. MergeBamAlignment considers and switches only existing primary and secondary designations. Therefore, it is critical that these were previously flagged.
![image]() A read with multiple alignment records may map to multiple loci or may be chimeric--that is, splits the alignment. It is possible for an aligner to produce multiple alignments as well as multiple primary alignments, e.g. in the case of a linear alignment set of split reads. When one alignment, or alignment set in the case of chimeric read records, is designated primary, others are designated either secondary or supplementary. Invoking the
A read with multiple alignment records may map to multiple loci or may be chimeric--that is, splits the alignment. It is possible for an aligner to produce multiple alignments as well as multiple primary alignments, e.g. in the case of a linear alignment set of split reads. When one alignment, or alignment set in the case of chimeric read records, is designated primary, others are designated either secondary or supplementary. Invoking the -M option, we had BWA mark the record with the longest aligning section of split reads as primary and all other records as secondary. MergeBamAlignment further adjusts this secondary designation and adds the read mapped in proper pair (0x2) and mate unmapped (0x8) flags. The tool then adjusts the strand orientation flag for a read (0x10) and it proper mate (0x20).
In the command, we change CLIP_ADAPTERS, MAX_INSERTIONS_OR_DELETIONS and PRIMARY_ALIGNMENT_STRATEGY values from default, and invoke other optional parameters. The path to the reference FASTA given by R should also contain the corresponding .dict sequence dictionary with the same prefix as the reference FASTA. It is imperative that both the uBAM and aligned BAM are both sorted by queryname.
Illustration of an intermediate step unused in workflow. See piped command.
java -Xmx16G -jar /path/picard.jar MergeBamAlignment \
R=/path/Homo_sapiens_assembly19.fasta \
UNMAPPED_BAM=6383_snippet_revertsam.bam \
ALIGNED_BAM=6483_snippet_bwa_mem.sam \ #accepts either SAM or BAM
O=6483_snippet_mergebamalignment.bam \
CREATE_INDEX=true \ #standard Picard option for coordinate-sorted outputs
ADD_MATE_CIGAR=true \ #default; adds MC tag
CLIP_ADAPTERS=false \ #changed from default
CLIP_OVERLAPPING_READS=true \ #default; soft-clips ends so mates do not extend past each other
INCLUDE_SECONDARY_ALIGNMENTS=true \ #default
MAX_INSERTIONS_OR_DELETIONS=-1 \ #changed to allow any number of insertions or deletions
PRIMARY_ALIGNMENT_STRATEGY=MostDistant \ #changed from default BestMapq
ATTRIBUTES_TO_RETAIN=XS \ #specify multiple times to retain tags starting with X, Y, or Z
TMP_DIR=/path/shlee #optional to process large files
This generates a coordinate-sorted and clean BAM, 6483_snippet_mergebamalignment.bam, and corresponding .bai index. These are ready for analyses starting with MarkDuplicates. The two bullet-point lists below describe changes to the resulting file. The first list gives general comments on select parameters and the second describes some of the notable changes to our example data.
Comments on select parameters
- Setting
PRIMARY_ALIGNMENT_STRATEGYto MostDistant marks primary alignments based on the alignment pair with the largest insert size. This strategy is based on the premise that of chimeric sections of a read aligning to consecutive regions, the alignment giving the largest insert size with the mate gives the most information.
- It may well be that alignments marked as secondary represent interesting biology, so we retain them with the
INCLUDE_SECONDARY_ALIGNMENTS parameter.
- Setting
MAX_INSERTIONS_OR_DELETIONS to -1 retains reads irregardless of the number of insertions and deletions. The default is 1.
- Because we leave the
ALIGNER_PROPER_PAIR_FLAGS parameter at the default false value, MergeBamAlignment will reassess and reassign proper pair designations made by the aligner. These are explained below using the example data.
ATTRIBUTES_TO_RETAIN is specified to carryover the XS tag from the alignment, which reports BWA-MEM's suboptimal alignment scores. My impression is that this is the next highest score for any alternative or additional alignments BWA considered, whether or not these additional alignments made it into the final aligned records. (IGV's BLAT feature allows you to search for additional sequence matches). For our tutorial data, this is the only additional unaccounted tag from the alignment. The XS tag in unnecessary for the Best Practices Workflow and is not retained by the Broad Genomics Platform's pipeline. We retain it here not only to illustrate that the tool carries over select alignment information only if asked, but also because I think it prudent. Given how compute intensive the alignment process is, the additional ~1% gain in the snippet file size seems a small price against having to rerun the alignment because we realize later that we want the tag.- Setting
CLIP_ADAPTERS to false leaves reads unclipped.
- By default the merged file is coordinate sorted. We set
CREATE_INDEX to true to additionally create the bai index.
- We need not invoke
PROGRAM options as BWA's program group information is sufficient and is retained in the merging.
- As a standalone tool, we would normally feed in a BAM file for
ALIGNED_BAM instead of the much larger SAM. We will be piping this step however and so need not add an extra conversion to BAM.
Description of changes to our example data
- MergeBamAlignment merges header information from the two sources that define read groups (@RG) and program groups (@PG) as well as reference contigs.
![image]() Tags are updated for our example data as shown in the table. The tool retains SA, MD, NM and AS tags from the alignment, given these are not present in the uBAM. The tool additionally adds UQ (the Phred likelihood of the segment), MC (mate CIGAR string) and MQ (mapping quality of the mate/next segment) tags if applicable. For unmapped reads (marked with an
Tags are updated for our example data as shown in the table. The tool retains SA, MD, NM and AS tags from the alignment, given these are not present in the uBAM. The tool additionally adds UQ (the Phred likelihood of the segment), MC (mate CIGAR string) and MQ (mapping quality of the mate/next segment) tags if applicable. For unmapped reads (marked with an * asterisk in column 6 of the SAM record), the tool removes AS and XS tags and assigns MC (if applicable), PG and RG tags. This is illustrated for example read H0164ALXX140820:2:1101:29704:6495 in the BWA-MEM section of this document.- Original base quality score restoration is illustrated in step 2.
The example below shows a read pair for which MergeBamAlignment adjusts multiple information fields, and these changes are described in the remaining bullet points.
- MergeBamAlignment changes hard-clipping to soft-clipping, e.g. 96H55M to 96S55M, and restores corresponding truncated sequences with the original full-length read sequence.
- The tool reorders the read records to reflect the chromosome and contig ordering in the header and the genomic coordinates for each.
- MergeBamAlignment's MostDistant
PRIMARY_ALIGNMENT_STRATEGY asks the tool to consider the best pair to mark as primary from the primary and secondary records. In this pair, one of the reads has two alignment loci, on contig hs37d5 and on chromosome 10. The two loci align 115 and 55 nucleotides, respectively, and the aligned sequences are identical by 55 bases. Flag values set by BWA-MEM indicate the contig hs37d5 record is primary and the shorter chromosome 10 record is secondary. For this chimeric read, MergeBamAlignment reassigns the chromosome 10 mapping as the primary alignment and the contig hs37d5 mapping as secondary (0x100 flag bit).
- In addition, MergeBamAlignment designates each record on chromosome 10 as read mapped in proper pair (0x2 flag bit) and the contig hs37d5 mapping as mate unmapped (0x8 flag bit). IGV's paired reads mode displays the two chromosome 10 mappings as a pair after these MergeBamAlignment adjustments.
- MergeBamAlignment adjusts read reverse strand (0x10 flag bit) and mate reverse strand (0x20 flag bit) flags consistent with changes to the proper pair designation. For our non-stranded DNA-Seq library alignments displayed in IGV, a read pointing rightward is in the forward direction (absence of 0x10 flag) and a read pointing leftward is in the reverse direction (flagged with 0x10). In a typical pair, where the rightward pointing read is to the left of the leftward pointing read, the left read will also have the mate reverse strand (0x20) flag.
Two distinct classes of mate unmapped read records are now present in our example file: (1) reads whose mates truly failed to map and are marked by an asterisk * in column 6 of the SAM record and (2) multimapping reads whose mates are in fact mapped but in a proper pair that excludes the particular read record. Each of these two classes of mate unmapped reads can contain multimapping reads that map to two or more locations.
Comparing 6483_snippet_bwa_mem.sam and 6483_snippet_mergebamalignment.bam, we see the number_unmapped reads_ remains the same at 1211, while the number of records with the mate unmapped flag increases by 1359, from 1276 to 2635. These now account for 0.951% of the 276,970 read records.
For 6483_snippet_mergebamalignment.bam, how many additional unique reads become mate unmapped?
After BWA-MEM alignment
![image]()
After MergeBamAlignment
![image]()
back to top
3D. Pipe SamToFastq, BWA-MEM and MergeBamAlignment to generate a clean BAM
![image]() We pipe the three tools described above to generate an aligned BAM file sorted by query name. In the piped command, the commands for the three processes are given together, separated by a vertical bar
We pipe the three tools described above to generate an aligned BAM file sorted by query name. In the piped command, the commands for the three processes are given together, separated by a vertical bar |. We also replace each intermediate output and input file name with a symbolic path to the system's output and input devices, here /dev/stdout and /dev/stdin, respectively. We need only provide the first input file and name the last output file.
Before using a piped command, we should ask UNIX to stop the piped command if any step of the pipe should error and also return to us the error messages. Type the following into your shell to set these UNIX options.
set -o pipefail
Overview of command structure
[SamToFastq] | [BWA-MEM] | [MergeBamAlignment]
Piped command
java -Xmx8G -jar /path/picard.jar SamToFastq \
I=6483_snippet_markilluminaadapters.bam \
FASTQ=/dev/stdout \
CLIPPING_ATTRIBUTE=XT CLIPPING_ACTION=2 INTERLEAVE=true NON_PF=true \
TMP_DIR=/path/shlee | \
/path/bwa mem -M -t 7 -p /path/Homo_sapiens_assembly19.fasta /dev/stdin | \
java -Xmx16G -jar /path/picard.jar MergeBamAlignment \
ALIGNED_BAM=/dev/stdin \
UNMAPPED_BAM=6383_snippet_revertsam.bam \
OUTPUT=6483_snippet_piped.bam \
R=/path/Homo_sapiens_assembly19.fasta CREATE_INDEX=true ADD_MATE_CIGAR=true \
CLIP_ADAPTERS=false CLIP_OVERLAPPING_READS=true \
INCLUDE_SECONDARY_ALIGNMENTS=true MAX_INSERTIONS_OR_DELETIONS=-1 \
PRIMARY_ALIGNMENT_STRATEGY=MostDistant ATTRIBUTES_TO_RETAIN=XS \
TMP_DIR=/path/shlee
The piped output file, 6483_snippet_piped.bam, is for all intensive purposes the same as 6483_snippet_mergebamalignment.bam, produced by running MergeBamAlignment separately without piping. However, the resulting files, as well as new runs of the workflow on the same data, have the potential to differ in small ways because each uses a different alignment instance.
How do these small differences arise?
Counting the number of mate unmapped reads shows that this number remains unchanged for the two described workflows. Two counts emitted at the end of the process updates, that also remain constant for these instances, are the number of alignment records and the number of unmapped reads.
INFO 2015-12-08 17:25:59 AbstractAlignmentMerger Wrote 275759 alignment records and 1211 unmapped reads.
back to top
Some final remarks
We have produced a clean BAM that is coordinate-sorted and indexed, in an efficient manner that minimizes processing time and storage needs. The file is ready for marking duplicates as outlined in Tutorial#2799. Additionally, we can now free up storage on our file system by deleting the original file we started with, the uBAM and the uBAMXT. We sleep well at night knowing that the clean BAM retains all original information.
We have two final comments (1) on multiplexed samples and (2) on fitting this workflow into a larger workflow.
For multiplexed samples, first perform the workflow steps on a file representing one sample and one lane. Then mark duplicates. Later, after some steps in the GATK's variant discovery workflow, and after aggregating files from the same sample from across lanes into a single file, mark duplicates again. These two marking steps ensure you find both optical and PCR duplicates.
For workflows that nestle this pipeline, consider additionally optimizing java jar's parameters for SamToFastq and MergeBamAlignment. For example, the following are the additional settings used by the Broad Genomics Platform in the piped command for very large data sets.
java -Dsamjdk.buffer_size=131072 -Dsamjdk.compression_level=1 -XX:GCTimeLimit=50 -XX:GCHeapFreeLimit=10 -Xmx128m -jar /path/picard.jar SamToFastq ...
java -Dsamjdk.buffer_size=131072 -Dsamjdk.use_async_io=true -Dsamjdk.compression_level=1 -XX:+UseStringCache -XX:GCTimeLimit=50 -XX:GCHeapFreeLimit=10 -Xmx5000m -jar /path/picard.jar MergeBamAlignment ...
I give my sincere thanks to Julian Hess, the GATK team and the Data Sciences and Data Engineering (DSDE) team members for all their help in writing this and related documents.
back to top
 If you are interested in emulating the methods used by the Broad Genomics Platform to pre-process your short read sequencing data, you have landed on the right page. The parsimonious operating procedures outlined in this three-step workflow both maximize data quality, storage and processing efficiency to produce a mapped and clean BAM. This clean BAM is ready for analysis workflows that start with MarkDuplicates.
If you are interested in emulating the methods used by the Broad Genomics Platform to pre-process your short read sequencing data, you have landed on the right page. The parsimonious operating procedures outlined in this three-step workflow both maximize data quality, storage and processing efficiency to produce a mapped and clean BAM. This clean BAM is ready for analysis workflows that start with MarkDuplicates.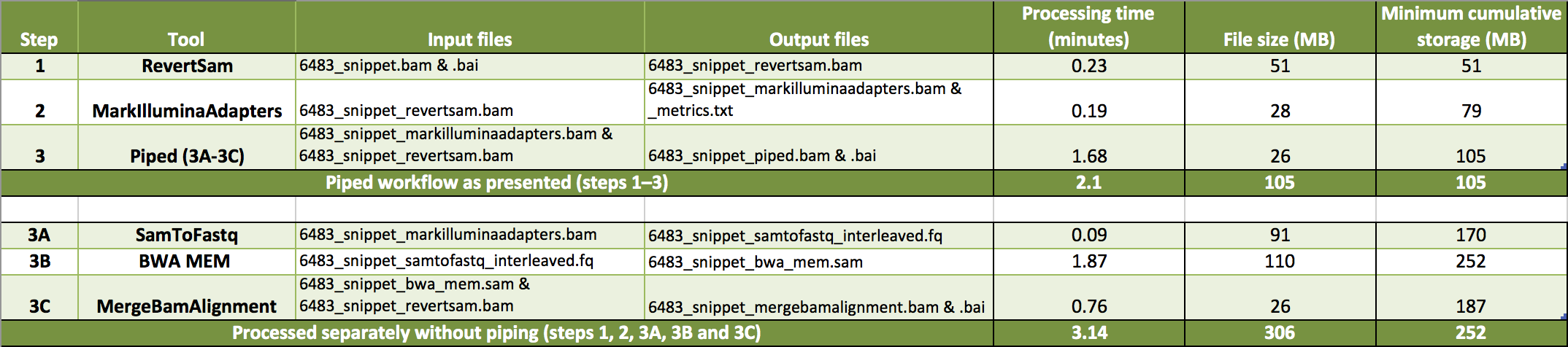









 A read with multiple alignment records may map to multiple loci or may be chimeric--that is, splits the alignment. It is possible for an aligner to produce multiple alignments as well as multiple primary alignments, e.g. in the case of a linear alignment set of split reads. When one alignment, or alignment set in the case of chimeric read records, is designated primary, others are designated either secondary or supplementary. Invoking the
A read with multiple alignment records may map to multiple loci or may be chimeric--that is, splits the alignment. It is possible for an aligner to produce multiple alignments as well as multiple primary alignments, e.g. in the case of a linear alignment set of split reads. When one alignment, or alignment set in the case of chimeric read records, is designated primary, others are designated either secondary or supplementary. Invoking the  Tags are updated for our example data as shown in the table. The tool retains SA, MD, NM and AS tags from the alignment, given these are not present in the uBAM. The tool additionally adds UQ (the Phred likelihood of the segment), MC (mate CIGAR string) and MQ (mapping quality of the mate/next segment) tags if applicable. For unmapped reads (marked with an
Tags are updated for our example data as shown in the table. The tool retains SA, MD, NM and AS tags from the alignment, given these are not present in the uBAM. The tool additionally adds UQ (the Phred likelihood of the segment), MC (mate CIGAR string) and MQ (mapping quality of the mate/next segment) tags if applicable. For unmapped reads (marked with an 

 We pipe the three tools described above to generate an aligned BAM file sorted by query name. In the piped command, the commands for the three processes are given together, separated by a
We pipe the three tools described above to generate an aligned BAM file sorted by query name. In the piped command, the commands for the three processes are given together, separated by a 




