How to identify duplicated genes in VCF file obtained after GATK pipeline?
Download small_exac_common_3_grch38.vcf.gz file
Hi, Is small_exac_common_3_grch38.vcf.gz publicly available?
I tried looking for this file in GATK bundle FTP site but could not find it.
Can you point me in the right direction to download this file?
Thanks for the help!
Mutect2 dont-use-soft-clipped-bases doesn't work properly
Hi guys,
I'm using Mutect2 from gatk version 4.1.2.0. to perform variant calling in tumor samples.
Since I'm using amplicon data I have clipped small portion of sequences at both end from the alignment before the variant calling.
Using Mutect2 with the option "--dont-use-soft-clipped-bases true" for the majority of the variants the depth is counted correctly.
However I get variants that is still counted as if it was not clipped.
I attach an example:
what I get without the --dont-use-soft-clipped-bases true option:
17 31235687 31235687 C A 17 31235687 . C A . . DP=61;ECNT=2;MBQ=39,39;MFRL=186,187;MMQ=36,33;MPOS=32;POPAF=7.30;TLOD=34.49 GT:AD:AF:DP:F1R2:F2R1:SB 0/1:48,13:0.222:61:14,13:34,0:34,14,0,13
what I get with the --dont-use-soft-clipped-bases true option:
17 31235687 31235687 C A 17 31235687 . C A . . DP=61;ECNT=2;MBQ=39,39;MFRL=186,187;MMQ=36,33;MPOS=32;POPAF=7.30;TLOD=34.49 GT:AD:AF:DP:F1R2:F2R1:SB 0/1:48,13:0.222:61:14,13:34,0:34,14,0,13
The total depth without considering the clipped reads should be 27 (14ref and 13 alt) but I still get 61, is there something that I can do to adjust the depth count?
Thanks for considering my request
'''
java -Xmx2g -jar /opt/gatk-4.1.2.0/gatk-package-4.1.2.0-local.jar Mutect2 --reference /data/REFERENCES/Genomes/hg38/human_g1k_v38.fasta --input ../MAPPING/SAMPLE.bam --dont-use-soft-clipped-bases true --output SAMPLE.mutect.variant
'''
SplitNCigarReads fails on unmapped reads in second pass (Illegal argument exception)
I'm trying to get RNA-seq data analysis-ready for variant calling according to Best Practices (RNAseq short variant discovery (SNPs + Indels)). SplitNCigarReads is failing and it seems like it might be a bug - I have pored over the previous steps and ValidateSamFile gives the bam file resulting from MarkDuplicates a clean bill of health. Also there have been a couple of similar questions on forums over the years but they were not resolved or closed as 'stale and probably fixed by now', i.e. https://github.com/bcbio/bcbio-nextgen/issues/2354 and https://gatkforums.broadinstitute.org/gatk/discussion/12547/splitncigarreads-fails-on-illegalargumentexception-contig-must-be-non-null-and-not-equal-to-and
Details:
Data - RNA-seq data downloaded from SRA, (SRR5511204); paired-end Illumina, 101bp reads. Alignment done with 2-pass STAR.
Platform: running locally from downloaded GATK version 4.1.4.0 on Ubuntu 16.04
Command (copied from program output):
```
java -Dsamjdk.use_async_io_read_samtools=false -Dsamjdk.use_async_io_write_samtools=true -Dsamjdk.use_async_io_write_tribble=false -Dsamjdk.compression_level=2 -jar /home/kate/Downloads/GATK/gatk-4.1.4.0/gatk-package-4.1.4.0-local.jar SplitNCigarReads -I marked_duplicates2.bam -O splitNcigarred.bam -R /home/kate/Databases/GRCh38/GRCh38.primary_assembly.genome.fa
```
Error (which seems to happen when program gets to unmapped reads in second pass):
```
java.lang.IllegalArgumentException: contig must be non-null and not equal to *, and start must be >= 1
at org.broadinstitute.hellbender.utils.read.SAMRecordToGATKReadAdapter.setMatePosition(SAMRecordToGATKReadAdapter.java:197)
at org.broadinstitute.hellbender.tools.walkers.rnaseq.OverhangFixingManager.setPredictedMateInformation(OverhangFixingManager.java:445)
at org.broadinstitute.hellbender.tools.walkers.rnaseq.SplitNCigarReads.splitNCigarRead(SplitNCigarReads.java:222)
at org.broadinstitute.hellbender.tools.walkers.rnaseq.SplitNCigarReads.secondPassApply(SplitNCigarReads.java:185)
at org.broadinstitute.hellbender.engine.TwoPassReadWalker.lambda$traverseReads$0(TwoPassReadWalker.java:72)
at java.util.stream.ForEachOps$ForEachOp$OfRef.accept(ForEachOps.java:184)
at java.util.stream.ReferencePipeline$3$1.accept(ReferencePipeline.java:193)
at java.util.stream.ReferencePipeline$2$1.accept(ReferencePipeline.java:175)
at java.util.stream.ReferencePipeline$3$1.accept(ReferencePipeline.java:193)
at java.util.Iterator.forEachRemaining(Iterator.java:116)
at java.util.Spliterators$IteratorSpliterator.forEachRemaining(Spliterators.java:1801)
at java.util.stream.AbstractPipeline.copyInto(AbstractPipeline.java:481)
at java.util.stream.AbstractPipeline.wrapAndCopyInto(AbstractPipeline.java:471)
at java.util.stream.ForEachOps$ForEachOp.evaluateSequential(ForEachOps.java:151)
at java.util.stream.ForEachOps$ForEachOp$OfRef.evaluateSequential(ForEachOps.java:174)
at java.util.stream.AbstractPipeline.evaluate(AbstractPipeline.java:234)
at java.util.stream.ReferencePipeline.forEach(ReferencePipeline.java:418)
at org.broadinstitute.hellbender.engine.TwoPassReadWalker.traverseReads(TwoPassReadWalker.java:70)
at org.broadinstitute.hellbender.engine.TwoPassReadWalker.traverse(TwoPassReadWalker.java:59)
at org.broadinstitute.hellbender.engine.GATKTool.doWork(GATKTool.java:1048)
at org.broadinstitute.hellbender.cmdline.CommandLineProgram.runTool(CommandLineProgram.java:139)
at org.broadinstitute.hellbender.cmdline.CommandLineProgram.instanceMainPostParseArgs(CommandLineProgram.java:191)
at org.broadinstitute.hellbender.cmdline.CommandLineProgram.instanceMain(CommandLineProgram.java:210)
at org.broadinstitute.hellbender.Main.runCommandLineProgram(Main.java:163)
at org.broadinstitute.hellbender.Main.mainEntry(Main.java:206)
at org.broadinstitute.hellbender.Main.main(Main.java:292)
```
Hope you can help because I'd really like to try variant calling!
Thanks :)
(How to) Install and use Conda for GATK4
Some tools in GATK4, like the gCNV pipeline and the new deep learning variant filtering tools, require extensive Python dependencies. To avoid having to worry about managing these dependencies, we recommend using the GATK4 docker container, which comes with everything pre-installed, as explained here. If you are running GATK4 on a server and/or cannot use the Docker image, we recommend using the Conda package manager as a backup solution. The Conda package manager comes with all the dependencies you need, so you do not need to install everything separately. Both Conda and Docker are intended to solve the same problem, but one of the big differences/benefits of Conda is that you can use Conda without having root access. Conda should be easy to install if you follow these steps.
1) Refer to the installation instructions from Conda. Choose the correct version/computer you need to download it for. You will have the option of downloading Anaconda or Miniconda. Conda provides documentation about the difference between Anaconda and Miniconda. We chose to use Miniconda for this tutorial because we just wanted to use the GATK conda environment and did not want to take up too much space on our computer. If you are not going to use Conda for anything other than GATK4, you might consider doing the same. If you choose to install Anaconda, you may have access to other bioinformatics packages that are helpful to you, and you won’t have to install each package you need. Follow the prompts to properly install the .pkg file. Make sure you choose the correct package for the version of Python you are using. For example, if you have Python 2.7 on your computer, choose the version specific to it.
2) Go to the directory where you have stored the GATK4 jars and the gatk wrapper script, and make sure gatkcondaenv.yml is present. Run
conda env create -n gatk -f gatkcondaenv.yml
source activate gatk
3) To check if your Conda environment is running properly, type conda list and you should see a list of packages installed.
gatkpythonpackages should be one of them.
4) You can also test out whether the new variant filtering tool (CNNScoreVariants) runs properly. If you run
python -c "import vqsr_cnn" the output should look like Using TensorFlow backend.. If you do not have the Conda environment configured correctly, you will get an error immediately saying ImportError: No module named vqsr_cnn.
5) If you later upgrade to a new version of GATK4, you will need to update the Conda configuration in the new GATK4 folder. If you simply overwrite the old GATK with the new one, you will get an error message saying “CondaValueError: prefix already exists: /anaconda2/envs/gatk”. For example, when I upgraded from GATK 4.0.1.2 to GATK 4.0.2.0, I simply ran (in my 4.0.2.0 folder)
source deactivate
conda env remove -n gatk
Then, follow Steps 2-4 again to re-install it.
Important
Do not confuse the above mentioned GATK conda environment setup with this bioconda gatk installation. The current version of the bioconda installation of GATK does not set up the conda environment used for the GATK python tools, so that must still be set up manually.
GT and AD
14 reads of the ALT and 4 that were not???? or something else
thank you
when will gatk support complex variant call like variants in gatk
anyone give some suggestion.
vardict is a choice, but it need to select the complex variant myself
Catching up with the times: GATK is moving to a new web home
TL,DR: In a few weeks, we're going to move the website and forum to a new online platform that will scale better with the needs of the research community. The website will still live at https://software.broadinstitute.org/gatk/ but there will be some important changes at the level of the user guide and the support forum in particular. Read on to get the lowdown on where this is coming from, where we're heading and how you can prepare for the upcoming migration.
A brief history
When GATK was first released around 2010, its documentation lived in a rather primitive wiki that was half-public, half-private, and almost entirely aimed at developers. The wiki was supplemented by a proto-forum, hosted by Get Satisfaction and run by Eric Banks, one of the original authors of GATK who has since risen to the lofty position of Senior Director in the Data Sciences Platform at the Broad (a heartwarming rags-to-riches story for another time). Despite being an absolutely lovely human being in person, Eric was notoriously mean to the unfortunate few who dared ask questions on the old forum. So, in 2012, I was hired to be a human filter for his snark, plus, you know, build a proper user guide. Something that would enable researchers to use GATK without needing to be physically in the room with the developers for the darn thing to work. Coming out of a wetlab microbiology postdoc, I was uniquely unqualified for the job, but that too is a story for another time... Point is, that's how a little over seven years ago, Roger the summer intern and I built and launched the GATK website, which included a more formally structured user guide and the community forum hosted by Vanilla Forums that we have been using ever since.
Our little hand-rolled artisanal website has had a good run, with over 20 million page views to date and about two to three thousand unique visitors on any given weekday. But it's time to face the facts: we've outgrown it. In that time, and especially since the release of GATK4 two years ago (OMG has it really been two years already), the toolkit has expanded dramatically. It currently includes more than 200 tools and multiple Best Practices pipelines covering the major variant classes, plus use cases like mitochondrial analysis. We're aware that many of you find it difficult to find the information you need in our sprawling documentation. And there's more new stuff coming out soon that we haven't yet had a chance to talk about... So it's clear we're going to need both a better structure and way more elbow room than the current system can support.
Long story short: it's time to reboot and rebuild
For the past few months, we've been crafting a new web home for GATK documentation and support. This time, instead of building a traditional website, we're using a customer service system called Zendesk that includes a knowledge base module for documentation and a community forum for Q&A. Part of our support team has already been using this system for the Terra helpdesk. Although the Terra knowledge base itself is still a work in progress, it’s been a positive experience so far. That gives us confidence that adopting Zendesk for GATK will help us improve the usability of the GATK documentation. We're also looking forward to being able to streamline our support operation by consolidating across the multiple software products and services offered by the Data Sciences Platform. That's good for everyone — not just those of you who use WDL and Terra as well as GATK — because if our support team spends less time wrangling different internal systems, they can spend more time improving the docs and answering your questions.
What's going to change?
Overall we're trying to minimize disruption but there will be a few important changes. Here's a breakdown of what you're most likely to care about.
The user guide will be organized a little differently and the search box will work better
We're taking this opportunity to update how the documentation content is organized to make it easier to find information. Hopefully this will be all upside, but if you get lost at any point, try the search box — it should work better in the new system.
Some links may break
This is perhaps the most important consequence of the fact that we're moving to a new content management system: all links to individual articles will change. We're going to set up an automated redirection system to map old URLs to the new ones, so that your bookmarks and links that people have previously posted online stay functional, but we can't guarantee that we'll be able to capture absolutely everything. We'll do our best to make the system handle missing content as gracefully as possible.
GATK3 documentation will be archived in GitHub
We're aware that the awkward coexistence of docs from the GATK3 era and the newer GATK4 versions is one of the major sources of confusion in the current user guide. We've wrestled with this problem and ultimately decided to move the GATK3 content to the documentation archive in the old GATK repository in GitHub, where versions 1.x through 3.x of the code live. This way the old content will always remain available for anyone still working with older versions of the software, yet it will be more clear that it only applies to those versions, and it will be out of the way of anyone using the current versions of the tools. And of course, we'll do our best to include all those articles in the redirect mapping to keep links functional.
You'll need a new account for the community forum
Unfortunately we're not able to move existing forum accounts to the new platform. So to ask questions, start discussions or add comments in the new Community forum, you’ll need to create a new account. The good news is that this new account will work for all the other products we support, like Terra and Cromwell/WDL. And if you already have a Terra community forum account, you’re all set.
The new system will support single sign-on (SSO) with Google, Microsoft, Twitter and Facebook.
Old forum discussions will eventually be taken offline
I'll admit it: this is the part that makes me hyperventilate a little. We have over 17,000 discussion threads in the "Ask a question" section of the forum, and it's just not feasible to migrate them all over to the new platform. Most of them are out of date anyway, referencing old tools and methods that we no longer recommend, command syntax that no longer works, and my favorite, bugs that no longer occur! But there is still plenty of useful information in there that's not in the docs, from explanations of weird errors to strategies for customizing methods for non-standard use cases. So we're going to keep the old forum online in read-only mode for the next few months, and during that time we'll comb through the most frequently visited threads to capture the good stuff and turn it into documentation articles. We're also open to suggestions if there are any discussions that you have found particularly useful in your own work.
However, at some point we're going to have to shut down the old forum. The plan right now is to shut it down on February 1st, 2020, but we'll re-evaluate that timing if we feel that we need more time for the knowledge capture process. Your opinion on this matters a lot to us, so don't hesitate to nominate threads that you think would be useful to preserve in the knowledge base. We also recommend you save any threads that are important to you personally as a PDF or HTML page on your computer just in case. If all else fails, the Internet Archive's "Way Back Machine" does preserve snapshots of the forum, so it's very likely that those old forum discussions will actually outlive us all.
Talk to us
Ultimately the purpose of these resources is to help you use GATK effectively in your work, so we'd really like to hear from you, especially if you have concerns about how any of this is going to affect how you normally use the documentation and forum. We're very open to making amendments based on your feedback, both before the migration happens but also during the months that follow. I have no doubt that as the dust settles and we put some mileage on the new platform, we'll see opportunities emerge to tweak it for the better. Don't be shy about volunteering your thoughts and suggestions!
Running HaplotypeCaller on a multi-lane sample
I am interested in running HaplotypeCaller for a multi-lane sample. What is the best way to do this? I followed the protocol here (https://software.broadinstitute.org/gatk/documentation/article?id=6057) which states to run MarkDuplicates and BQSR before HaplotypeCaller, which I did. I fed the new BAM file into HaplotypeCaller but get an error with a corrupt gVCF file. Am I meant to merge the read groups into a single read group before using HaplotypeCaller?
Unable to filter VCF file using VariantFiltration for GATK 3.7.0
1) I am using GATK 3.7 version for VariantFiltration step. I have generated vcf file using HaplotypeCaller, run GenotypeGVCF
followed by SNP and indel recalibration, snpEff and VariantAnnotator before running this VariantFiltration step.
All the steps have been run using GATK 3.7.0 version.
It gives me following error when I try to run this command
GenomeAnalysisTK -T VariantFiltration \
-R genome.fasta \
-V input.ann.vcf \
--filterExpression "GQ < 20.0" --filterName "GQ" \
--filterExpression "VQSLOD <= 0" --filterName "VQSLOD" \
-o trial.vcf \
Error:
##### ERROR MESSAGE: Invalid argument value '<' at position 8.
##### ERROR Invalid argument value '20.0' at position 9.
##### ERROR Invalid argument value '<=' at position 16.
##### ERROR Invalid argument value '0' at position 17.
Secondly I would like to filter variants in Subtelomeric regions based on intervals file as follows
Chr1 1 27336 SubtelomericRepeat
Chr1 27337 92900 SubtelomericHypervariable
Chr1 92901 457931 Core
Chr1 457932 460311 Centromere
Chr1 460312 575900 Core
Please
(How to) Map and clean up short read sequence data efficiently
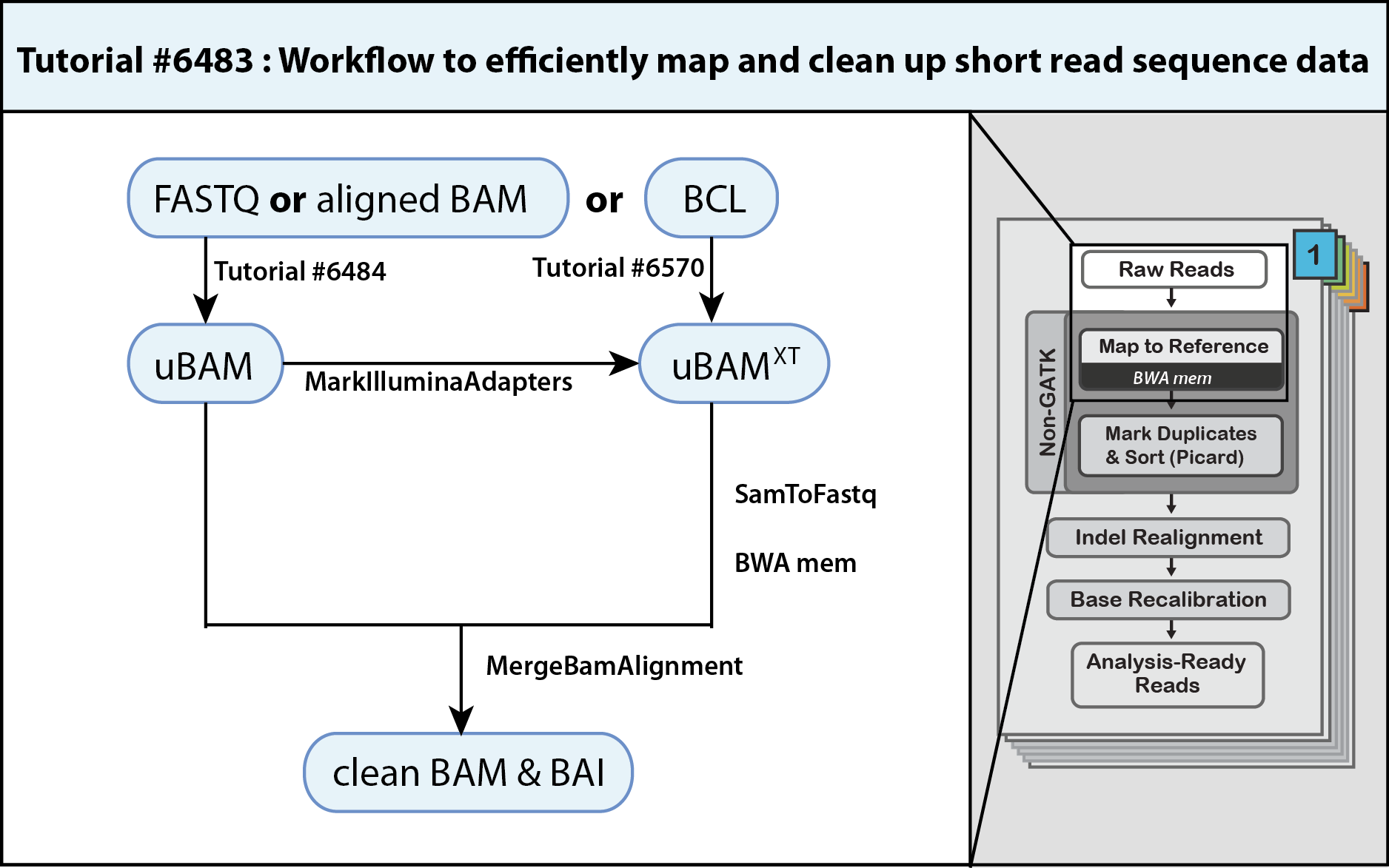 If you are interested in emulating the methods used by the Broad Genomics Platform to pre-process your short read sequencing data, you have landed on the right page. The parsimonious operating procedures outlined in this three-step workflow both maximize data quality, storage and processing efficiency to produce a mapped and clean BAM. This clean BAM is ready for analysis workflows that start with MarkDuplicates.
If you are interested in emulating the methods used by the Broad Genomics Platform to pre-process your short read sequencing data, you have landed on the right page. The parsimonious operating procedures outlined in this three-step workflow both maximize data quality, storage and processing efficiency to produce a mapped and clean BAM. This clean BAM is ready for analysis workflows that start with MarkDuplicates.
Since your sequencing data could be in a number of formats, the first step of this workflow refers you to specific methods to generate a compatible unmapped BAM (uBAM, Tutorial#6484) or (uBAMXT, Tutorial#6570 coming soon). Not all unmapped BAMs are equal and these methods emphasize cleaning up prior meta information while giving you the opportunity to assign proper read group fields. The second step of the workflow has you marking adapter sequences, e.g. arising from read-through of short inserts, using MarkIlluminaAdapters such that they contribute minimally to alignments and allow the aligner to map otherwise unmappable reads. The third step pipes three processes to produce the final BAM. Piping SamToFastq, BWA-MEM and MergeBamAlignment saves time and allows you to bypass storage of larger intermediate FASTQ and SAM files. In particular, MergeBamAlignment merges defined information from the aligned SAM with that of the uBAM to conserve read data, and importantly, it generates additional meta information and unifies meta data. The resulting clean BAM is coordinate sorted, indexed.
The workflow reflects a lossless operating procedure that retains original sequencing read information within the final BAM file such that data is amenable to reversion and analysis by different means. These practices make scaling up and long-term storage efficient, as one needs only keep the final BAM file.
Geraldine_VdAuwera points out that there are many different ways of correctly preprocessing HTS data for variant discovery and ours is only one approach. So keep this in mind.
We present this workflow using real data from a public sample. The original data file, called Solexa-272222, is large at 150 GB. The file contains 151 bp paired PCR-free reads giving 30x coverage of a human whole genome sample referred to as NA12878. The entire sample library was sequenced in a single flow cell lane and thereby assigns all the reads the same read group ID. The example commands work both on this large file and on smaller files containing a subset of the reads, collectively referred to as snippet. NA12878 has a variant in exon 5 of the CYP2C19 gene, on the portion of chromosome 10 covered by the snippet, resulting in a nonfunctional protein. Consistent with GATK's recommendation of using the most up-to-date tools, for the given example results, with the exception of BWA, we used the most current versions of tools as of their testing (September to December 2015). We provide illustrative example results, some of which were derived from processing the original large file and some of which show intermediate stages skipped by this workflow.
Download example snippet data to follow along the tutorial.
We welcome feedback. Share your suggestions in the Comments section at the bottom of this page.
Jump to a section
- Generate an unmapped BAM from FASTQ, aligned BAM or BCL
- Mark adapter sequences using MarkIlluminaAdapters
- Align reads with BWA-MEM and merge with uBAM using MergeBamAlignment
A. Convert BAM to FASTQ and discount adapter sequences using SamToFastq
B. Align reads and flag secondary hits using BWA-MEM
C. Restore altered data and apply & adjust meta information using MergeBamAlignment
D. Pipe SamToFastq, BWA-MEM and MergeBamAlignment to generate a clean BAM
Tools involved
- MarkIlluminaAdapters
- Unix pipelines
- SamToFastq
- BWA-MEM (Li 2013 reference; Li 2014 benchmarks; homepage; manual)
- MergeBamAlignment
Prerequisites
- Installed Picard tools
- Installed GATK tools
- Installed BWA
- Reference genome
- Illumina or similar tech DNA sequence reads file containing data corresponding to one read group ID. That is, the file contains data from one sample and from one flow cell lane.
Download example data
- To download the reference, open ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/2.8/b37/ in your browser. Leave the password field blank. Download the following three files (~860 MB) to the same folder:
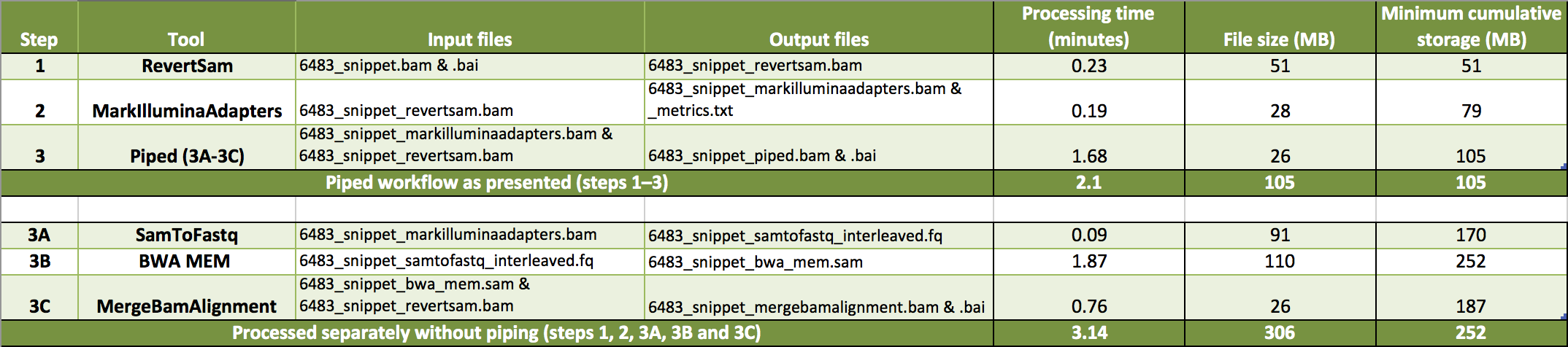
human_g1k_v37_decoy.fasta.gz,.fasta.fai.gz, and.dict.gz. This same reference is available to load in IGV. - I divided the example data into two tarballs: tutorial_6483_piped.tar.gz contains the files for the piped process and tutorial_6483_intermediate_files.tar.gz contains the intermediate files produced by running each process independently. The data contain reads originally aligning to a one Mbp genomic interval (10:96,000,000-97,000,000) of GRCh37. The table shows the steps of the workflow, corresponding input and output example data files and approximate minutes and disk space needed to process each step. Additionally, we tabulate the time and minimum storage needed to complete the workflow as presented (piped) or without piping.
Related resources
- See this tutorial to add or replace read groups or coordinate-sort and index a BAM.
- See this tutorial for basic instructions on using the Integrative Genomics Viewer (IGV).
- For collecting alignment summary metrics, see CollectAlignmentSummaryMetrics, CollectWgsMetrics and CollectInsertSizeMetrics. See Picard for metrics definitions.
- See SAM flags to interpret SAM flag values.
- Tutorial#2799 gives an example command to mark duplicates.
Other notes
- When transforming data files, we stick to using Picard tools over other tools to avoid subtle incompatibilities.
For large files, (1) use the Java
-Xmxsetting and (2) set the environmental variableTMP_DIRfor a temporary directory.java -Xmx8G -jar /path/picard.jar MarkIlluminaAdapters \ TMP_DIR=/path/shleeIn the command, the
-Xmx8GJava option caps the maximum heap size, or memory usage, to eight gigabytes. The path given byTMP_DIRpoints the tool to scratch space that it can use. These options allow the tool to run without slowing down as well as run without causing an out of memory error. The-Xmxsettings we provide here are more than sufficient for most cases. For GATK, 4G is standard, while for Picard less is needed. Some tools, e.g. MarkDuplicates, may require more. These options can be omitted for small files such as the example data and the equivalent command is as follows.java -jar /path/picard.jar MarkIlluminaAdaptersTo find a system's default maximum heap size, type
java -XX:+PrintFlagsFinal -version, and look forMaxHeapSize. Note that any setting beyond available memory spills to storage and slows a system down. If multithreading, increase memory proportionately to the number of threads. e.g. if 1G is the minimum required for one thread, then use 2G for two threads.When I call default options within a command, follow suit to ensure the same results.
1. Generate an unmapped BAM from FASTQ, aligned BAM or BCL
If you have raw reads data in BAM format with appropriately assigned read group fields, then you can start with step 2. Namely, besides differentiating samples, the read group ID should differentiate factors contributing to technical batch effects, i.e. flow cell lane. If not, you need to reassign read group fields. This dictionary post describes factors to consider and this post and this post provide some strategic advice on handling multiplexed data.
- See this tutorial to add or replace read groups.
If your reads are mapped, or in BCL or FASTQ format, then generate an unmapped BAM according to the following instructions.
- To convert FASTQ or revert aligned BAM files, follow directions in Tutorial#6484. The resulting uBAM needs to have its adapter sequences marked as outlined in the next step (step 2).
- To convert an Illumina Base Call files (BCL) use IlluminaBasecallsToSam. The tool marks adapter sequences at the same time. The resulting uBAMXT has adapter sequences marked with the XT tag so you can skip step 2 of this workflow and go directly to step 3. The corresponding Tutorial#6570 is coming soon.
See if you can revert
6483_snippet.bam, containing 279,534 aligned reads, to the unmapped6383_snippet_revertsam.bam, containing 275,546 reads.
2. Mark adapter sequences using MarkIlluminaAdapters
MarkIlluminaAdapters adds the XT tag to a read record to mark the 5' start position of the specified adapter sequence and produces a metrics file. Some of the marked adapters come from concatenated adapters that randomly arise from the primordial soup that is a PCR reaction. Others represent read-through to 3' adapter ends of reads and arise from insert sizes that are shorter than the read length. In some instances read-though can affect the majority of reads in a sample, e.g. in Nextera library samples over-titrated with transposomes, and render these reads unmappable by certain aligners. Tools such as SamToFastq use the XT tag in various ways to effectively remove adapter sequence contribution to read alignment and alignment scoring metrics. Depending on your library preparation, insert size distribution and read length, expect varying amounts of such marked reads.
java -Xmx8G -jar /path/picard.jar MarkIlluminaAdapters \
I=6483_snippet_revertsam.bam \
O=6483_snippet_markilluminaadapters.bam \
M=6483_snippet_markilluminaadapters_metrics.txt \ #naming required
TMP_DIR=/path/shlee #optional to process large files
This produces two files. (1) The metrics file, 6483_snippet_markilluminaadapters_metrics.txt bins the number of tagged adapter bases versus the number of reads. (2) The 6483_snippet_markilluminaadapters.bam file is identical to the input BAM, 6483_snippet_revertsam.bam, except reads with adapter sequences will be marked with a tag in XT:i:# format, where # denotes the 5' starting position of the adapter sequence. At least six bases are required to mark a sequence. Reads without adapter sequence remain untagged.
- By default, the tool uses Illumina adapter sequences. This is sufficient for our example data.
- Adjust the default standard Illumina adapter sequences to any adapter sequence using the
FIVE_PRIME_ADAPTERandTHREE_PRIME_ADAPTERparameters. To clear and add new adapter sequences first setADAPTERSto 'null' then specify each sequence with the parameter.
We plot the metrics data that is in GATKReport file format using RStudio, and as you can see, marked bases vary in size up to the full length of reads.


Do you get the same number of marked reads?
6483_snippetmarks 448 reads (0.16%) with XT, while the originalSolexa-272222marks 3,236,552 reads (0.39%).
Below, we show a read pair marked with the XT tag by MarkIlluminaAdapters. The insert region sequences for the reads overlap by a length corresponding approximately to the XT tag value. For XT:i:20, the majority of the read is adapter sequence. The same read pair is shown after SamToFastq transformation, where adapter sequence base quality scores have been set to 2 (# symbol), and after MergeBamAlignment, which restores original base quality scores.
Unmapped uBAM (step 1)

After MarkIlluminaAdapters (step 2)

After SamToFastq (step 3)

After MergeBamAlignment (step 3)

3. Align reads with BWA-MEM and merge with uBAM using MergeBamAlignment
This step actually pipes three processes, performed by three different tools. Our tutorial example files are small enough to easily view, manipulate and store, so any difference in piped or independent processing will be negligible. For larger data, however, using Unix pipelines can add up to significant savings in processing time and storage.
Not all tools are amenable to piping and piping the wrong tools or wrong format can result in anomalous data.
The three tools we pipe are SamToFastq, BWA-MEM and MergeBamAlignment. By piping these we bypass storage of larger intermediate FASTQ and SAM files. We additionally save time by eliminating the need for the processor to read in and write out data for two of the processes, as piping retains data in the processor's input-output (I/O) device for the next process.
To make the information more digestible, we will first talk about each tool separately. At the end of the section, we provide the piped command.
3A. Convert BAM to FASTQ and discount adapter sequences using SamToFastq
Picard's SamToFastq takes read identifiers, read sequences, and base quality scores to write a Sanger FASTQ format file. We use additional options to effectively remove previously marked adapter sequences, in this example marked with an XT tag. By specifying CLIPPING_ATTRIBUTE=XT and CLIPPING_ACTION=2, SamToFastq changes the quality scores of bases marked by XT to two--a rather low score in the Phred scale. This effectively removes the adapter portion of sequences from contributing to downstream read alignment and alignment scoring metrics.
Illustration of an intermediate step unused in workflow. See piped command.
java -Xmx8G -jar /path/picard.jar SamToFastq \
I=6483_snippet_markilluminaadapters.bam \
FASTQ=6483_snippet_samtofastq_interleaved.fq \
CLIPPING_ATTRIBUTE=XT \
CLIPPING_ACTION=2 \
INTERLEAVE=true \
NON_PF=true \
TMP_DIR=/path/shlee #optional to process large files
This produces a FASTQ file in which all extant meta data, i.e. read group information, alignment information, flags and tags are purged. What remains are the read query names prefaced with the @ symbol, read sequences and read base quality scores.
- For our paired reads example file we set SamToFastq's
INTERLEAVEto true. During the conversion to FASTQ format, the query name of the reads in a pair are marked with /1 or /2 and paired reads are retained in the same FASTQ file. BWA aligner accepts interleaved FASTQ files given the-poption. - We change the
NON_PF, akaINCLUDE_NON_PF_READS, option from default to true. SamToFastq will then retain reads marked by what some consider an archaic 0x200 flag bit that denotes reads that do not pass quality controls, aka reads failing platform or vendor quality checks. Our tutorial data do not contain such reads and we call out this option for illustration only. - Other CLIPPING_ACTION options include (1) X to hard-clip, (2) N to change bases to Ns or (3) another number to change the base qualities of those positions to the given value.
3B. Align reads and flag secondary hits using BWA-MEM
In this workflow, alignment is the most compute intensive and will take the longest time. GATK's variant discovery workflow recommends Burrows-Wheeler Aligner's maximal exact matches (BWA-MEM) algorithm (Li 2013 reference; Li 2014 benchmarks; homepage; manual). BWA-MEM is suitable for aligning high-quality long reads ranging from 70 bp to 1 Mbp against a large reference genome such as the human genome.
- Aligning our
snippetreads against either a portion or the whole genome is not equivalent to aligning our originalSolexa-272222file, merging and taking a newslicefrom the same genomic interval. - For the tutorial, we use BWA v 0.7.7.r441, the same aligner used by the Broad Genomics Platform as of this writing (9/2015).
- As mentioned, alignment is a compute intensive process. For faster processing, use a reference genome with decoy sequences, also called a decoy genome. For example, the Broad's Genomics Platform uses an Hg19/GRCh37 reference sequence that includes Ebstein-Barr virus (EBV) sequence to soak up reads that fail to align to the human reference that the aligner would otherwise spend an inordinate amount of time trying to align as split reads. GATK's resource bundle provides a standard decoy genome from the 1000 Genomes Project.
BWA alignment requires an indexed reference genome file. Indexing is specific to algorithms. To index the human genome for BWA, we apply BWA's
indexfunction on the reference genome file, e.g.human_g1k_v37_decoy.fasta. This produces five index files with the extensionsamb,ann,bwt,pacandsa.bwa index -a bwtsw human_g1k_v37_decoy.fasta
The example command below aligns our example data against the GRCh37 genome. The tool automatically locates the index files within the same folder as the reference FASTA file.
Illustration of an intermediate step unused in workflow. See piped command.
/path/bwa mem -M -t 7 -p /path/human_g1k_v37_decoy.fasta \
6483_snippet_samtofastq_interleaved.fq > 6483_snippet_bwa_mem.sam
This command takes the FASTQ file, 6483_snippet_samtofastq_interleaved.fq, and produces an aligned SAM format file, 6483_snippet_unthreaded_bwa_mem.sam, containing read alignment information, an automatically generated program group record and reads sorted in the same order as the input FASTQ file. Aligner-assigned alignment information, flag and tag values reflect each read's or split read segment's best sequence match and does not take into consideration whether pairs are mapped optimally or if a mate is unmapped. Added tags include the aligner-specific XS tag that marks secondary alignment scores in XS:i:# format. This tag is given for each read even when the score is zero and even for unmapped reads. The program group record (@PG) in the header gives the program group ID, group name, group version and recapitulates the given command. Reads are sorted by query name. For the given version of BWA, the aligned file is in SAM format even if given a BAM extension.
Does the aligned file contain read group information?
We invoke three options in the command.
-Mto flag shorter split hits as secondary.
This is optional for Picard compatibility as MarkDuplicates can directly process BWA's alignment, whether or not the alignment marks secondary hits. However, if we want MergeBamAlignment to reassign proper pair alignments, to generate data comparable to that produced by the Broad Genomics Platform, then we must mark secondary alignments.-pto indicate the given file contains interleaved paired reads.-tfollowed by a number for the number of processor threads to use concurrently. Here we use seven threads which is one less than the total threads available on my Mac laptap. Check your server or system's total number of threads with the following command provided by KateN.getconf _NPROCESSORS_ONLN
In the example data, all of the 1211 unmapped reads each have an asterisk (*) in column 6 of the SAM record, where a read typically records its CIGAR string. The asterisk represents that the CIGAR string is unavailable. The several asterisked reads I examined are recorded as mapping exactly to the same location as their _mapped_ mates but with MAPQ of zero. Additionally, the asterisked reads had varying noticeable amounts of low base qualities, e.g. strings of #s, that corresponded to original base quality calls and not those changed by SamToFastq. This accounting by BWA allows these pairs to always list together, even when the reads are coordinate-sorted, and leaves a pointer to the genomic mapping of the mate of the unmapped read. For the example read pair shown below, comparing sequences shows no apparent overlap, with the highest identity at 72% over 25 nts.
After MarkIlluminaAdapters (step 2)

After BWA-MEM (step 3)

After MergeBamAlignment (step 3)

3C. Restore altered data and apply & adjust meta information using MergeBamAlignment
MergeBamAlignment is a beast of a tool, so its introduction is longer. It does more than is implied by its name. Explaining these features requires I fill you in on some background.
Broadly, the tool merges defined information from the unmapped BAM (uBAM, step 1) with that of the aligned BAM (step 3) to conserve read data, e.g. original read information and base quality scores. The tool also generates additional meta information based on the information generated by the aligner, which may alter aligner-generated designations, e.g. mate information and secondary alignment flags. The tool then makes adjustments so that all meta information is congruent, e.g. read and mate strand information based on proper mate designations. We ascribe the resulting BAM as clean.
Specifically, the aligned BAM generated in step 3 lacks read group information and certain tags--the UQ (Phred likelihood of the segment), MC (CIGAR string for mate) and MQ (mapping quality of mate) tags. It has hard-clipped sequences from split reads and altered base qualities. The reads also have what some call mapping artifacts but what are really just features we should not expect from our aligner. For example, the meta information so far does not consider whether pairs are optimally mapped and whether a mate is unmapped (in reality or for accounting purposes). Depending on these assignments, MergeBamAlignment adjusts the read and read mate strand orientations for reads in a proper pair. Finally, the alignment records are sorted by query name. We would like to fix all of these issues before taking our data to a variant discovery workflow.
Enter MergeBamAlignment. As the tool name implies, MergeBamAlignment applies read group information from the uBAM and retains the program group information from the aligned BAM. In restoring original sequences, the tool adjusts CIGAR strings from hard-clipped to soft-clipped. If the alignment file is missing reads present in the unaligned file, then these are retained as unmapped records. Additionally, MergeBamAlignment evaluates primary alignment designations according to a user-specified strategy, e.g. for optimal mate pair mapping, and changes secondary alignment and mate unmapped flags based on its calculations. Additional for desired congruency. I will soon explain these and additional changes in more detail and show a read record to illustrate.
Consider what
PRIMARY_ALIGNMENT_STRATEGYoption best suits your samples. MergeBamAlignment applies this strategy to a read for which the aligner has provided more than one primary alignment, and for which one is designated primary by virtue of another record being marked secondary. MergeBamAlignment considers and switches only existing primary and secondary designations. Therefore, it is critical that these were previously flagged.
 A read with multiple alignment records may map to multiple loci or may be chimeric--that is, splits the alignment. It is possible for an aligner to produce multiple alignments as well as multiple primary alignments, e.g. in the case of a linear alignment set of split reads. When one alignment, or alignment set in the case of chimeric read records, is designated primary, others are designated either secondary or supplementary. Invoking the
A read with multiple alignment records may map to multiple loci or may be chimeric--that is, splits the alignment. It is possible for an aligner to produce multiple alignments as well as multiple primary alignments, e.g. in the case of a linear alignment set of split reads. When one alignment, or alignment set in the case of chimeric read records, is designated primary, others are designated either secondary or supplementary. Invoking the -M option, we had BWA mark the record with the longest aligning section of split reads as primary and all other records as secondary. MergeBamAlignment further adjusts this secondary designation and adds the read mapped in proper pair (0x2) and mate unmapped (0x8) flags. The tool then adjusts the strand orientation flag for a read (0x10) and it proper mate (0x20).
In the command, we change CLIP_ADAPTERS, MAX_INSERTIONS_OR_DELETIONS and PRIMARY_ALIGNMENT_STRATEGY values from default, and invoke other optional parameters. The path to the reference FASTA given by R should also contain the corresponding .dict sequence dictionary with the same prefix as the reference FASTA. It is imperative that both the uBAM and aligned BAM are both sorted by queryname.
Illustration of an intermediate step unused in workflow. See piped command.
java -Xmx16G -jar /path/picard.jar MergeBamAlignment \
R=/path/Homo_sapiens_assembly19.fasta \
UNMAPPED_BAM=6383_snippet_revertsam.bam \
ALIGNED_BAM=6483_snippet_bwa_mem.sam \ #accepts either SAM or BAM
O=6483_snippet_mergebamalignment.bam \
CREATE_INDEX=true \ #standard Picard option for coordinate-sorted outputs
ADD_MATE_CIGAR=true \ #default; adds MC tag
CLIP_ADAPTERS=false \ #changed from default
CLIP_OVERLAPPING_READS=true \ #default; soft-clips ends so mates do not extend past each other
INCLUDE_SECONDARY_ALIGNMENTS=true \ #default
MAX_INSERTIONS_OR_DELETIONS=-1 \ #changed to allow any number of insertions or deletions
PRIMARY_ALIGNMENT_STRATEGY=MostDistant \ #changed from default BestMapq
ATTRIBUTES_TO_RETAIN=XS \ #specify multiple times to retain tags starting with X, Y, or Z
TMP_DIR=/path/shlee #optional to process large files
This generates a coordinate-sorted and clean BAM, 6483_snippet_mergebamalignment.bam, and corresponding .bai index. These are ready for analyses starting with MarkDuplicates. The two bullet-point lists below describe changes to the resulting file. The first list gives general comments on select parameters and the second describes some of the notable changes to our example data.
Comments on select parameters
- Setting
PRIMARY_ALIGNMENT_STRATEGYto MostDistant marks primary alignments based on the alignment pair with the largest insert size. This strategy is based on the premise that of chimeric sections of a read aligning to consecutive regions, the alignment giving the largest insert size with the mate gives the most information. - It may well be that alignments marked as secondary represent interesting biology, so we retain them with the
INCLUDE_SECONDARY_ALIGNMENTSparameter. - Setting
MAX_INSERTIONS_OR_DELETIONSto -1 retains reads irregardless of the number of insertions and deletions. The default is 1. - Because we leave the
ALIGNER_PROPER_PAIR_FLAGSparameter at the default false value, MergeBamAlignment will reassess and reassign proper pair designations made by the aligner. These are explained below using the example data. ATTRIBUTES_TO_RETAINis specified to carryover the XS tag from the alignment, which reports BWA-MEM's suboptimal alignment scores. My impression is that this is the next highest score for any alternative or additional alignments BWA considered, whether or not these additional alignments made it into the final aligned records. (IGV's BLAT feature allows you to search for additional sequence matches). For our tutorial data, this is the only additional unaccounted tag from the alignment. The XS tag in unnecessary for the Best Practices Workflow and is not retained by the Broad Genomics Platform's pipeline. We retain it here not only to illustrate that the tool carries over select alignment information only if asked, but also because I think it prudent. Given how compute intensive the alignment process is, the additional ~1% gain in thesnippetfile size seems a small price against having to rerun the alignment because we realize later that we want the tag.- Setting
CLIP_ADAPTERSto false leaves reads unclipped. - By default the merged file is coordinate sorted. We set
CREATE_INDEXto true to additionally create thebaiindex. - We need not invoke
PROGRAMoptions as BWA's program group information is sufficient and is retained in the merging. - As a standalone tool, we would normally feed in a BAM file for
ALIGNED_BAMinstead of the much larger SAM. We will be piping this step however and so need not add an extra conversion to BAM.
Description of changes to our example data
- MergeBamAlignment merges header information from the two sources that define read groups (@RG) and program groups (@PG) as well as reference contigs.
![image]() Tags are updated for our example data as shown in the table. The tool retains SA, MD, NM and AS tags from the alignment, given these are not present in the uBAM. The tool additionally adds UQ (the Phred likelihood of the segment), MC (mate CIGAR string) and MQ (mapping quality of the mate/next segment) tags if applicable. For unmapped reads (marked with an
Tags are updated for our example data as shown in the table. The tool retains SA, MD, NM and AS tags from the alignment, given these are not present in the uBAM. The tool additionally adds UQ (the Phred likelihood of the segment), MC (mate CIGAR string) and MQ (mapping quality of the mate/next segment) tags if applicable. For unmapped reads (marked with an *asterisk in column 6 of the SAM record), the tool removes AS and XS tags and assigns MC (if applicable), PG and RG tags. This is illustrated for example readH0164ALXX140820:2:1101:29704:6495in the BWA-MEM section of this document.- Original base quality score restoration is illustrated in step 2.
The example below shows a read pair for which MergeBamAlignment adjusts multiple information fields, and these changes are described in the remaining bullet points.
- MergeBamAlignment changes hard-clipping to soft-clipping, e.g. 96H55M to 96S55M, and restores corresponding truncated sequences with the original full-length read sequence.
- The tool reorders the read records to reflect the chromosome and contig ordering in the header and the genomic coordinates for each.
- MergeBamAlignment's MostDistant
PRIMARY_ALIGNMENT_STRATEGYasks the tool to consider the best pair to mark as primary from the primary and secondary records. In this pair, one of the reads has two alignment loci, on contig hs37d5 and on chromosome 10. The two loci align 115 and 55 nucleotides, respectively, and the aligned sequences are identical by 55 bases. Flag values set by BWA-MEM indicate the contig hs37d5 record is primary and the shorter chromosome 10 record is secondary. For this chimeric read, MergeBamAlignment reassigns the chromosome 10 mapping as the primary alignment and the contig hs37d5 mapping as secondary (0x100 flag bit). - In addition, MergeBamAlignment designates each record on chromosome 10 as read mapped in proper pair (0x2 flag bit) and the contig hs37d5 mapping as mate unmapped (0x8 flag bit). IGV's paired reads mode displays the two chromosome 10 mappings as a pair after these MergeBamAlignment adjustments.
- MergeBamAlignment adjusts read reverse strand (0x10 flag bit) and mate reverse strand (0x20 flag bit) flags consistent with changes to the proper pair designation. For our non-stranded DNA-Seq library alignments displayed in IGV, a read pointing rightward is in the forward direction (absence of 0x10 flag) and a read pointing leftward is in the reverse direction (flagged with 0x10). In a typical pair, where the rightward pointing read is to the left of the leftward pointing read, the left read will also have the mate reverse strand (0x20) flag.
Two distinct classes of mate unmapped read records are now present in our example file: (1) reads whose mates truly failed to map and are marked by an asterisk
*in column 6 of the SAM record and (2) multimapping reads whose mates are in fact mapped but in a proper pair that excludes the particular read record. Each of these two classes of mate unmapped reads can contain multimapping reads that map to two or more locations.
Comparing 6483_snippet_bwa_mem.sam and 6483_snippet_mergebamalignment.bam, we see the number_unmapped reads_ remains the same at 1211, while the number of records with the mate unmapped flag increases by 1359, from 1276 to 2635. These now account for 0.951% of the 276,970 read records.
For
6483_snippet_mergebamalignment.bam, how many additional unique reads become mate unmapped?
After BWA-MEM alignment

After MergeBamAlignment

3D. Pipe SamToFastq, BWA-MEM and MergeBamAlignment to generate a clean BAM
 We pipe the three tools described above to generate an aligned BAM file sorted by query name. In the piped command, the commands for the three processes are given together, separated by a vertical bar
We pipe the three tools described above to generate an aligned BAM file sorted by query name. In the piped command, the commands for the three processes are given together, separated by a vertical bar |. We also replace each intermediate output and input file name with a symbolic path to the system's output and input devices, here /dev/stdout and /dev/stdin, respectively. We need only provide the first input file and name the last output file.
Before using a piped command, we should ask UNIX to stop the piped command if any step of the pipe should error and also return to us the error messages. Type the following into your shell to set these UNIX options.
set -o pipefail
Overview of command structure
[SamToFastq] | [BWA-MEM] | [MergeBamAlignment]
Piped command
java -Xmx8G -jar /path/picard.jar SamToFastq \
I=6483_snippet_markilluminaadapters.bam \
FASTQ=/dev/stdout \
CLIPPING_ATTRIBUTE=XT CLIPPING_ACTION=2 INTERLEAVE=true NON_PF=true \
TMP_DIR=/path/shlee | \
/path/bwa mem -M -t 7 -p /path/Homo_sapiens_assembly19.fasta /dev/stdin | \
java -Xmx16G -jar /path/picard.jar MergeBamAlignment \
ALIGNED_BAM=/dev/stdin \
UNMAPPED_BAM=6383_snippet_revertsam.bam \
OUTPUT=6483_snippet_piped.bam \
R=/path/Homo_sapiens_assembly19.fasta CREATE_INDEX=true ADD_MATE_CIGAR=true \
CLIP_ADAPTERS=false CLIP_OVERLAPPING_READS=true \
INCLUDE_SECONDARY_ALIGNMENTS=true MAX_INSERTIONS_OR_DELETIONS=-1 \
PRIMARY_ALIGNMENT_STRATEGY=MostDistant ATTRIBUTES_TO_RETAIN=XS \
TMP_DIR=/path/shlee
The piped output file, 6483_snippet_piped.bam, is for all intensive purposes the same as 6483_snippet_mergebamalignment.bam, produced by running MergeBamAlignment separately without piping. However, the resulting files, as well as new runs of the workflow on the same data, have the potential to differ in small ways because each uses a different alignment instance.
How do these small differences arise?
Counting the number of mate unmapped reads shows that this number remains unchanged for the two described workflows. Two counts emitted at the end of the process updates, that also remain constant for these instances, are the number of alignment records and the number of unmapped reads.
INFO 2015-12-08 17:25:59 AbstractAlignmentMerger Wrote 275759 alignment records and 1211 unmapped reads.
Some final remarks
We have produced a clean BAM that is coordinate-sorted and indexed, in an efficient manner that minimizes processing time and storage needs. The file is ready for marking duplicates as outlined in Tutorial#2799. Additionally, we can now free up storage on our file system by deleting the original file we started with, the uBAM and the uBAMXT. We sleep well at night knowing that the clean BAM retains all original information.
We have two final comments (1) on multiplexed samples and (2) on fitting this workflow into a larger workflow.
For multiplexed samples, first perform the workflow steps on a file representing one sample and one lane. Then mark duplicates. Later, after some steps in the GATK's variant discovery workflow, and after aggregating files from the same sample from across lanes into a single file, mark duplicates again. These two marking steps ensure you find both optical and PCR duplicates.
For workflows that nestle this pipeline, consider additionally optimizing java jar's parameters for SamToFastq and MergeBamAlignment. For example, the following are the additional settings used by the Broad Genomics Platform in the piped command for very large data sets.
java -Dsamjdk.buffer_size=131072 -Dsamjdk.compression_level=1 -XX:GCTimeLimit=50 -XX:GCHeapFreeLimit=10 -Xmx128m -jar /path/picard.jar SamToFastq ...
java -Dsamjdk.buffer_size=131072 -Dsamjdk.use_async_io=true -Dsamjdk.compression_level=1 -XX:+UseStringCache -XX:GCTimeLimit=50 -XX:GCHeapFreeLimit=10 -Xmx5000m -jar /path/picard.jar MergeBamAlignment ...
I give my sincere thanks to Julian Hess, the GATK team and the Data Sciences and Data Engineering (DSDE) team members for all their help in writing this and related documents.
GATK Determine Contig Ploidy Error: ValueError: invalid literal for int() with base 10: '0 PLOIDY_P
Hi,
I am running GATK4 Germline CNV pipeline on 203 WES samples, and have gotten the following error during the Determine Contig Ploidy step:
15:29:38.431 DEBUG ScriptExecutor - Executing:
15:29:38.431 DEBUG ScriptExecutor - python
15:29:38.431 DEBUG ScriptExecutor - /tmp/cohort_determine_ploidy_and_depth.5886214504352644800.py
15:29:38.431 DEBUG ScriptExecutor - --sample_coverage_metadata=/tmp/samples-by-coverage-per-contig8026198779202597096.tsv
15:29:38.431 DEBUG ScriptExecutor - --output_calls_path=/mnt/data/smb_share/Mandal_project/PCa_WES/mcdowell#Bailey-Wilson_NF_AAPC/SampleBams/ploidy-calls
15:29:38.431 DEBUG ScriptExecutor - --mapping_error_rate=1.000000e-02
15:29:38.431 DEBUG ScriptExecutor - --psi_s_scale=1.000000e-04
15:29:38.431 DEBUG ScriptExecutor - --mean_bias_sd=1.000000e-02
15:29:38.431 DEBUG ScriptExecutor - --psi_j_scale=1.000000e-03
15:29:38.432 DEBUG ScriptExecutor - --learning_rate=5.000000e-02
15:29:38.432 DEBUG ScriptExecutor - --adamax_beta1=9.000000e-01
15:29:38.432 DEBUG ScriptExecutor - --adamax_beta2=9.990000e-01
15:29:38.432 DEBUG ScriptExecutor - --log_emission_samples_per_round=2000
15:29:38.432 DEBUG ScriptExecutor - --log_emission_sampling_rounds=100
15:29:38.432 DEBUG ScriptExecutor - --log_emission_sampling_median_rel_error=5.000000e-04
15:29:38.432 DEBUG ScriptExecutor - --max_advi_iter_first_epoch=1000
15:29:38.432 DEBUG ScriptExecutor - --max_advi_iter_subsequent_epochs=1000
15:29:38.432 DEBUG ScriptExecutor - --min_training_epochs=20
15:29:38.432 DEBUG ScriptExecutor - --max_training_epochs=100
15:29:38.432 DEBUG ScriptExecutor - --initial_temperature=2.000000e+00
15:29:38.432 DEBUG ScriptExecutor - --num_thermal_advi_iters=5000
15:29:38.432 DEBUG ScriptExecutor - --convergence_snr_averaging_window=5000
15:29:38.432 DEBUG ScriptExecutor - --convergence_snr_trigger_threshold=1.000000e-01
15:29:38.432 DEBUG ScriptExecutor - --convergence_snr_countdown_window=10
15:29:38.432 DEBUG ScriptExecutor - --max_calling_iters=1
15:29:38.432 DEBUG ScriptExecutor - --caller_update_convergence_threshold=1.000000e-03
15:29:38.432 DEBUG ScriptExecutor - --caller_internal_admixing_rate=7.500000e-01
15:29:38.432 DEBUG ScriptExecutor - --caller_external_admixing_rate=7.500000e-01
15:29:38.432 DEBUG ScriptExecutor - --disable_caller=false
15:29:38.432 DEBUG ScriptExecutor - --disable_sampler=false
15:29:38.432 DEBUG ScriptExecutor - --disable_annealing=false
15:29:38.432 DEBUG ScriptExecutor - --interval_list=/tmp/intervals8489974735940571592.tsv
15:29:38.432 DEBUG ScriptExecutor - --contig_ploidy_prior_table=/mnt/data/smb_share/Mandal_project/PCa_WES/mcdowell#Bailey-Wilson_NF_AAPC/SampleBams/contigPloidyPriorsTable4.tsv
15:29:38.432 DEBUG ScriptExecutor - --output_model_path=/mnt/data/smb_share/Mandal_project/PCa_WES/mcdowell#Bailey-Wilson_NF_AAPC/SampleBams/ploidy-model
Traceback (most recent call last):
File "/tmp/cohort_determine_ploidy_and_depth.5886214504352644800.py", line 79, in <module>
args.contig_ploidy_prior_table)
File "/usr/miniconda3/envs/gatk/lib/python3.6/site-packages/gcnvkernel/io/io_ploidy.py", line 190, in get_contig_ploidy_prior_map_from_tsv_file
ploidy_values = [int(column[len(io_consts.ploidy_prior_prefix):]) for column in columns[1:]]
File "/usr/miniconda3/envs/gatk/lib/python3.6/site-packages/gcnvkernel/io/io_ploidy.py", line 190, in <listcomp>
ploidy_values = [int(column[len(io_consts.ploidy_prior_prefix):]) for column in columns[1:]]
ValueError: invalid literal for int() with base 10: '0 PLOIDY_PRIOR_1 PLOIDY_PRIOR_2 PLOIDY_PRIOR_3
15:29:52.389 DEBUG ScriptExecutor - Result: 1
15:29:52.390 INFO DetermineGermlineContigPloidy - Shutting down engine
[June 26, 2019 3:29:52 PM CDT] org.broadinstitute.hellbender.tools.copynumber.DetermineGermlineContigPloidy done. Elapsed time: 5.24 minutes.
Runtime.totalMemory()=6116343808
org.broadinstitute.hellbender.utils.python.PythonScriptExecutorException:
python exited with 1
Command Line: python /tmp/cohort_determine_ploidy_and_depth.5886214504352644800.py --sample_coverage_metadata=/tmp/samples-by-coverage-per-contig8026198779202597096.tsv --output_calls_path=/mnt/data/smb_share/Mandal_project/PCa_WES/mcdowell#Bailey-Wilson_NF_AAPC/SampleBams/ploidy-calls --mapping_error_rate=1.000000e-02 --psi_s_scale=1.000000e-04 --mean_bias_sd=1.000000e-02 --psi_j_scale=1.000000e-03 --learning_rate=5.000000e-02 --adamax_beta1=9.000000e-01 --adamax_beta2=9.990000e-01 --log_emission_samples_per_round=2000 --log_emission_sampling_rounds=100 --log_emission_sampling_median_rel_error=5.000000e-04 --max_advi_iter_first_epoch=1000 --max_advi_iter_subsequent_epochs=1000 --min_training_epochs=20 --max_training_epochs=100 --initial_temperature=2.000000e+00 --num_thermal_advi_iters=5000 --convergence_snr_averaging_window=5000 --convergence_snr_trigger_threshold=1.000000e-01 --convergence_snr_countdown_window=10 --max_calling_iters=1 --caller_update_convergence_threshold=1.000000e-03 --caller_internal_admixing_rate=7.500000e-01 --caller_external_admixing_rate=7.500000e-01 --disable_caller=false --disable_sampler=false --disable_annealing=false --interval_list=/tmp/intervals8489974735940571592.tsv --contig_ploidy_prior_table=/mnt/data/smb_share/Mandal_project/PCa_WES/mcdowell#Bailey-Wilson_NF_AAPC/SampleBams/contigPloidyPriorsTable4.tsv --output_model_path=/mnt/data/smb_share/Mandal_project/PCa_WES/mcdowell#Bailey-Wilson_NF_AAPC/SampleBams/ploidy-model
at org.broadinstitute.hellbender.utils.python.PythonExecutorBase.getScriptException(PythonExecutorBase.java:75)
at org.broadinstitute.hellbender.utils.runtime.ScriptExecutor.executeCuratedArgs(ScriptExecutor.java:126)
at org.broadinstitute.hellbender.utils.python.PythonScriptExecutor.executeArgs(PythonScriptExecutor.java:170)
at org.broadinstitute.hellbender.utils.python.PythonScriptExecutor.executeScript(PythonScriptExecutor.java:151)
at org.broadinstitute.hellbender.utils.python.PythonScriptExecutor.executeScript(PythonScriptExecutor.java:121)
at org.broadinstitute.hellbender.tools.copynumber.DetermineGermlineContigPloidy.executeDeterminePloidyAndDepthPythonScript(DetermineGermlineContigPloidy.java:411)
at org.broadinstitute.hellbender.tools.copynumber.DetermineGermlineContigPloidy.doWork(DetermineGermlineContigPloidy.java:288)
at org.broadinstitute.hellbender.cmdline.CommandLineProgram.runTool(CommandLineProgram.java:138)
at org.broadinstitute.hellbender.cmdline.CommandLineProgram.instanceMainPostParseArgs(CommandLineProgram.java:191)
at org.broadinstitute.hellbender.cmdline.CommandLineProgram.instanceMain(CommandLineProgram.java:210)
at org.broadinstitute.hellbender.Main.runCommandLineProgram(Main.java:162)
at org.broadinstitute.hellbender.Main.mainEntry(Main.java:205)
at org.broadinstitute.hellbender.Main.main(Main.java:291)
We are using this contig ploidy table below:
CONTIG_NAME PLOIDY_PRIOR_0 PLOIDY_PRIOR_1 PLOIDY_PRIOR_2 PLOIDY_PRIOR_3
chr1 0.01 0.02 0.95 0.02
chr2 0.01 0.02 0.95 0.02
chr3 0.01 0.02 0.95 0.02
chr4 0.01 0.02 0.95 0.02
chr5 0.01 0.02 0.95 0.02
chr6 0.01 0.02 0.95 0.02
chr7 0.01 0.02 0.95 0.02
chr8 0.01 0.02 0.95 0.02
chr9 0.01 0.02 0.95 0.02
chr10 0.01 0.02 0.95 0.02
chr11 0.01 0.02 0.95 0.02
chr12 0.01 0.02 0.95 0.02
chr13 0.01 0.02 0.95 0.02
chr14 0.01 0.02 0.95 0.02
chr15 0.01 0.02 0.95 0.02
chr16 0.01 0.02 0.95 0.02
chr17 0.01 0.02 0.95 0.02
chr18 0.01 0.02 0.95 0.02
chr19 0.01 0.02 0.95 0.02
chr20 0.01 0.02 0.95 0.02
chr21 0.01 0.02 0.95 0.02
chr22 0.01 0.02 0.95 0.02
chrX 0.01 0.49 0.48 0.02
chrY 0.49 0.49 0.02 0
Any help would be appreciated.
Thanks,
Tarun
Enter the DRAGEN-GATK
It's a beautiful early autumn day in New England, with small patches of vibrant reds and yellows in the foliage just hinting at the fiery displays to come. Perfect weather for me to de-lurk and bring you some news! (I promise it's not GATK5)
The long and short of it (but mostly the short) is that we've started collaborating with the DRAGEN team at Illumina, led by Rami Mehio, to improve GATK tools and pipelines. There's a press release if you want the official announcement, or you can read on to get the long version from the GATK team's perspective.
If you're not familiar with DRAGEN, the name stands for Dynamic Read Analysis for GENomics and refers to a secondary analysis platform originally created by a company called Edico Genome, which was acquired by Illumina last year. The DRAGEN team became widely known for making genomic data processing insanely fast on special hardware, but they're not just a speed shop. They have top-notch computational biology expertise: when they reimplemented GATK tools like HaplotypeCaller in DRAGEN, they made some clever tweaks that improved the scientific accuracy of the results. They've done this for other tools as well, and they've also developed their own novel algorithms for other use cases.
That alone is already a big motivation for us to team up with them: they have great ideas for improving our tools and pipelines, and they're willing to share them. Works for us! Then there's the bigger picture of what this means for the kind of research we are working to enable. Both of our teams feel pretty strongly that as the amount of genomic data generation snowballs, particularly in the biomedical field, it's really important to ensure that the results of different studies can be cross-analyzed. For that to be possible, we need to standardize secondary analysis as much as possible to minimize batch effects. We believe that by working together to consolidate our methods and pipeline development efforts, we can remove a major source of heterogeneity in the ecosystem.
So what does that mean in practice?
Rest assured GATK itself is still going to be GATK, developed by our team at the Broad and released under the same BSD-3 open-source license you know and love. Any improvements that the DRAGEN team contributes to GATK tools will be integrated into the GATK codebase under the same BSD-3 license.
Beyond code improvements to GATK itself, there will also be some changes to the composition of the Best Practices pipelines. For example, we're going to replace BWA with the DRAGEN aligner, which is quite a bit faster, in our DNA pre-processing pipelines (full details and benchmarking results to follow). To reflect the collaborative nature of the work, any pipelines we co-develop with the DRAGEN team will be named DRAGEN-GATK Best Practices.
All the software involved in the DRAGEN-GATK pipelines will be fully open source and available in Github, including a new open source version of the DRAGEN aligner, and we'll continue to publish WDL workflows for every pipeline in Github and in Terra workspaces. Importantly, it will all still be runnable on normal hardware, whether you're doing your work on a local server, on-premises HPC or in the cloud. We'll also continue to provide free support for all GATK tools and pipelines, and as part of that we're going to work with the DRAGEN team to make sure we can provide the same level of high quality support for the tools that they provide.

The DRAGEN team also plans to produce a hardware-accelerated version of any DRAGEN-GATK Best Practices pipeline that we co-develop, which Illumina will offer on the commercial DRAGEN system. We won't touch that work at all (it's not our jam), but we will run comparative evaluations to validate that the hardware-accelerated version of any given pipeline produces results that are functionally equivalent to the "universal" open source software version. To be clear, it won't be just a rubber-stamp approval; we're highly motivated to make sure that the pipeline implementations are functionally equivalent because our colleagues in the Broad’s Genomics Platform are planning to switch some of the Broad's production pipelines to the DRAGEN hardware version for projects where speed is a critical factor.
On that note, what I personally find the most exciting about this partnership is that going forward, everyone in the research community will be able to take advantage of the best ideas from both our teams regardless of whether they want the "regular" software or a hardware-accelerated version. You could even switch between the two within the course of a project and still be able to cross-analyze the outputs. Over the years, I've had to tell a lot of people "sorry, you're going to have to reprocess everything with the same pipeline" so this feels like a huge step in the right direction.
Okay, this sounds great -- so when will the improved tools and pipelines be available?
We're already actively working on porting over improvements from the DRAGEN team, so if you follow the GATK repository on Github you should start seeing relevant commits and pull requests any day now. Barring any unforeseen complications, the tool improvements should roll out into regular GATK releases over the next couple of months, and we expect to release the first full DRAGEN-GATK pipeline (for germline short variants) in the first quarter of 2020. We'll post updates here on the blog about how it's going and what you can expect to see as the code rolls in and the release calendar firms up.
In the meantime, don't hesitate to reach out to us if you have any questions that aren't addressed here or in the press release. Note that if you're going to be at the ASHG meeting in Houston later this month, Angel Pizarro and I will be talking about this collaboration at the Illumina Informatics Summit that precedes the conference on Tuesday Oct 15, and I will be available at the Broad Genomics booth in the exhibit hall at ASHG itself on Wednesday Oct 16 if you'd like to discuss this in person. I hope to see a lot of you there!
Errors in CombineGVCF
I have converted already generated bam files (old project) in to GVCF using GATK4. Bam files were generated using GATK3 (older version). I tried to Combine 50 GVCF files. I encountered an error of malformed GVCF file. I searched about this. To deal with this I followed already given solutions - regenerate idx files, validate variants and regenerate GVCF again. idx creation didn't help. ValidateVariants failed for all my GVCFs (not sure why, though test run of 3 files was successful). To deal with this later, I omitted this file to joint rest 49. Again, I got an error -
01:08:08.917 INFO ProgressMeter - 1:16621897 18.2 119613000 6580730.3
01:08:18.918 INFO ProgressMeter - 1:16754369 18.3 120875000 6589731.2
01:08:26.432 INFO CombineGVCFs - Shutting down engine
[November 22, 2019 1:08:26 AM MST] org.broadinstitute.hellbender.tools.walkers.CombineGVCFs done. Elapsed time: 23.99 minutes.
Runtime.totalMemory()=95565643776
htsjdk.tribble.TribbleException$InternalCodecException: The following invalid GT allele index was encountered in the file: <NON_REF>
I performed interval padding and interval merging as suggested but didn't help.
Commands I used :
Haplotype Caller -
gatk HaplotypeCaller --java-options "-Xmx8G -XX:+UseParallelGC -XX:ParallelGCThreads=4" -R ../reference/GRCh37.fa -I test.bam -O test.raw.snps.indels.g.vcf -L b37_wgs_calling_regions.v1.list -ERC GVCF
CombineGVCF
gatk --java-options "-Xmx150g" CombineGVCFs -V test.list -R ../reference/GRCh37.fa -O Combine_49.g.vcf -ip 50 -imr ALL
Another error I encountered is missing INFO in VCF header for different set of GVCF.
Though as a test I performed on 3 GVCF files, they combined perfectly. Please help to resolve these.With so many errors in this step, I am thinking of using old UnifiedGenotyper.
Thank you
Ankita
Where can I find dbsnp_144.hg38.vcf.gz
I'm installing an application that uses files from the GATK resource bundle. I found all of the needed files at https://console.cloud.google.com/storage/browser/genomics-public-data/resources/broad/hg38/v0 , with the exception of one, dbsnp_144.hg38.vcf.gz.
It is no longer available at ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/ . Where can I find this file?
Mutect2 4.1.4.0 stats file with a negative number
Hello, I've just adapted my pipeline to the new filtering strategies, while looking at the files I noticed that for a WGS run I obtained a stats file with a negative number:
[egrassi@occam biodiversa]>cat mutect/CRC1307LMO.vcf.gz.stats
statistic value
callable -1.538687311E9
Looking around about the meaning of the number I found https://gatkforums.broadinstitute.org/gatk/discussion/24496/regenerating-mutect2-stats-file, so I'm wondering if I should be worried by having a negative number of callable sites 
What's more puzzling is that FilterMutectCalls after ran without any error.
Before running mutect I used the usual best practices pipeline, then:
gatk Mutect2 -tumor CRC1307LMO -R /archive/home/egrassi/bit/task/annotations/dataset/gnomad/GRCh38.d1.vd1.fa -I align/realigned_CRC1307LMO.bam -O mutect/CRC1307LMO.vcf.gz --germline-resource /archive/home/egrassi/bit/task/annotations/dataset/gnomad/af-only-gnomad.hg38.vcf.gz --f1r2-tar-gz mutect/CRC1307LMO_f1r2.tar.gz --independent-mates 2> mutect/CRC1307LMO.vcf.gz.log
gatk CalculateContamination -I mutect/CRC1307LMO.pileup.table -O mutect/CRC1307LMO.contamination.table --tumor-segmentation mutect/CRC1307LMO.tum.seg 2> mutect/CRC1307LMO.contamination.table.log
gatk LearnReadOrientationModel -I mutect/CRC1307LMO_f1r2.tar.gz -O mutect/CRC1307LMO_read-orientation-model.tar.gz 2> mutect/CRC1307LMO_read-orientation-model.tar.gz.log
gatk FilterMutectCalls -V mutect/CRC1307LMO.vcf.gz -O mutect/CRC1307LMO.filtered.vcf.gz -R /archive/home/egrassi/bit/task/annotations/dataset/gnomad/GRCh38.d1.vd1.fa --stats mutect/CRC1307LMO.vcf.gz.stats --contamination-table mutect/CRC1307LMO.contamination.table --tumor-segmentation=mutect/CRC1307LMO.tum.seg --filtering-stats mutect/CRC1307LMO_filtering_stats.tsv --ob-priors mutect/CRC1307LMO_read-orientation-model.tar.gz 2> mutect/CRC1307LMO_filtering_stats.tsv.log
GT with AD data meaning
I understand that the AD provides the "unfiltered" allele depth, i.e. the number of reads that support each of the reported alleles and that the GT field provides the genotype of the sample and that "0" is the REF allele and "1" represents the first ALT allele. (1/1 is a homozygous alternate sample--0/0 is a homozygous reference sample)
My question was seeking understanding of the combination of the two pieces of "coupled" data: GT and AD
If the sample is homozygous alternate (1/1) and the AD is 14,4;how do I interpret those 2 pieces of data when they are for the same "position" (on the same line or on the same record or for the same position)??
Again I apologize for not making my question clear, and
Which part of mym QD plot is the homozygous peak?
Hi there,
Just a quick question, which I think may be of use to people with similarly...squiffy...plots! I've plotted out QD values v Density so as to inform the hard filtering process but I'm having difficulty discerning the expected peaks for heterozygous calls and homozygous calls, as described at https://software.broadinstitute.org/gatk/guide/article?id=6925. As you can see from the attached plot, there is a peak at the lower values (or is it a shoulder?), a tiny bump and then a major peak, but then just a shoulder on the other side of the peak. As filtering effectively (and stringently) is key to my study, I'd like to know what each peak and shoulder represents before I take the plunge if anyone can make an educated guess, please?
Many thanks,
Ian
Got error of java.lang.IllegalArgumentException: Invalid interval. Contig:81 start:0 end:69 with Fil
gatk --java-options "-Xmx500m" FilterAlignmentArtifacts \
-R human_g1k_v37_decoy.fasta \
-V sample.filtered.vcf \ #come form Mutect2 After FilterMutectCalls
-I sample.sorted.out.bam \ #come from Mutect2
--bwa-mem-index-image hg37_reference.fasta.img \
-O sample.filtered.rm_artifacts.vcf.gz
So I really want to know how to fix this problem, I'v search this problem in forum but failed to find related themes.Please Help Me with best appreciation for GATK team
*** Error in `java': munmap_chunk(): invalid pointer: 0x00002ab8193572c0 ***running FilterMutectCall
*** Error in `java': munmap_chunk(): invalid pointer: 0x00002ab8193572c0 ***
======= Backtrace: =========
/lib64/libc.so.6(+0x78466)[0x2ab8142ce466]
/tmp/libgkl_smithwaterman7245711869685548422.so(_Z19runSWOnePairBT_avx2iiiiPhS_iiaPcPs+0x338)[0x2ab83420efa8]
/tmp/libgkl_smithwaterman7245711869685548422.so(Java_com_intel_gkl_smithwaterman_IntelSmithWaterman_alignNative+0xd8)[0x2ab83420ebf8]
[0x2ab81cf9e152]
======= Memory map: ========
00400000-00401000 r-xp 00000000 00:12 12740986699 /ifs/TJPROJ3/DISEASE/share/Software/Java/jdk1.8.0_51/bin/java
00600000-00601000 rw-p 00000000 00:12 12740986699 /ifs/TJPROJ3/DISEASE/share/Software/Java/jdk1.8.0_51/bin/java
006c6000-006e7000 rw-p 00000000 00:00 0 [heap]
e0c00000-f5580000 rw-p 00000000 00:00 0
f5580000-f5980000 ---p 00000000 00:00 0
f5980000-ff780000 rw-p 00000000 00:00 0
ff780000-100000000 ---p 00000000 00:00 0
100000000-1005c0000 rw-p 00000000 00:00 0
1005c0000-140000000 ---p 00000000 00:00 0
2ab8139fa000-2ab813a1c000 r-xp 00000000 08:01 655779 /lib64/ld-2.17.so
....
So,what is the possible problem with this? Please give me some help
 Tags are updated for our example data as shown in the table. The tool retains SA, MD, NM and AS tags from the alignment, given these are not present in the uBAM. The tool additionally adds UQ (the Phred likelihood of the segment), MC (mate CIGAR string) and MQ (mapping quality of the mate/next segment) tags if applicable. For unmapped reads (marked with an
Tags are updated for our example data as shown in the table. The tool retains SA, MD, NM and AS tags from the alignment, given these are not present in the uBAM. The tool additionally adds UQ (the Phred likelihood of the segment), MC (mate CIGAR string) and MQ (mapping quality of the mate/next segment) tags if applicable. For unmapped reads (marked with an